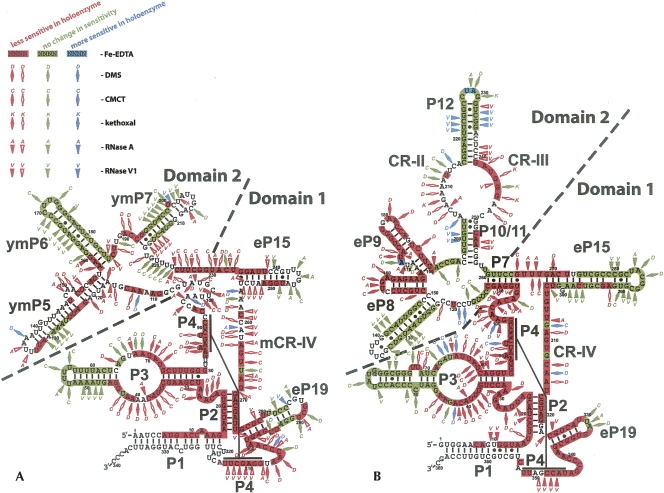
FIGURE 2.
Regions of RNase MRP (A) and RNase P (B) RNAs affected by protein binding in the holoenzyme. Red markings signify higher level of protection in the holoenzyme versus deproteinated RNA; green markings indicate no change in sensitivity upon deproteination; blue markings indicate lower sensitivity upon deproteination. Highlighting of nucleotides describes the results of the Fe-EDTA footprinting assays (indicative of the exposure of the RNA backbone). Diamonds describe the results of chemical probing (indicative of the exposure of the bases; D- DMS modifications; C- CMCT modifications; K- kethoxal modifications). Triangles describe the results of enzymatic probing (A- RNase A; V- RNase V1). Empty red diamonds or triangles correspond to weaker (1.5× to 4.5×) protection; filled red diamonds or triangles correspond to stronger (5× and up) protection. The diagrams and nomenclature of the structural elements are generally based on the work of Frank et al. (2000), Li et al. (2002), and Walker and Avis (2004) with minor corrections (Materials and Methods).