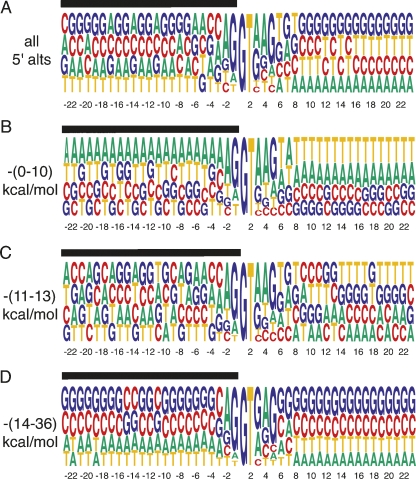
FIGURE 2.
Alignment of alternative 5′ splice sites +23 to −23 nt from the splice site. (A) A nucleotide frequency matrix constructed from all 5′ alternative splice sites. (B) Alternative 5′ splice sites with free energies between (0–10)-kcal/mol favor low stability RNA structures as indicated by the preference for A and T. (C) 5′ splice sites with folding free energies between (11–13)-kcal/mol shows no discernable sequence preference. (D) Alternative 5′ splice sites with free energies between (14–36)-kcal/mol favor the formation of strong RNA secondary structures as indicated by the preference for G and C. The x-axis denotes the distance from the splice site, the black bar the exon.