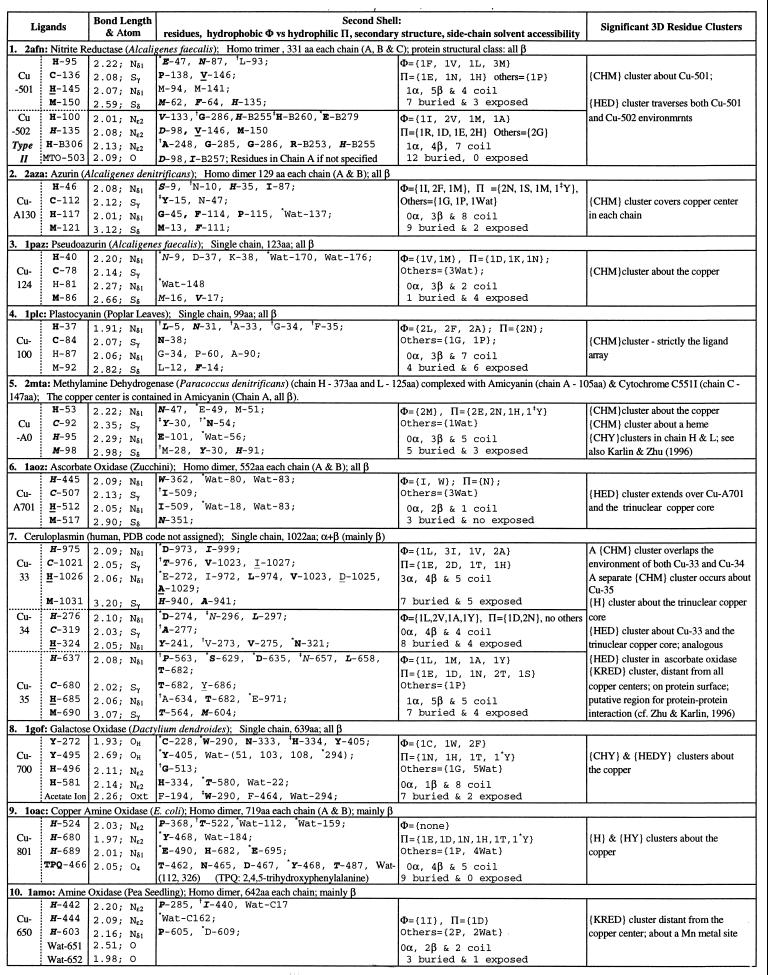
Table 1.
Single copper ions in protein 3D structures
Copper centers 1–7 are classified type I (blue copper); copper centers 8–10 are type II. A ligand residue or a residue in the second shell is shown by the one-letter amino acid code followed by chain identifier and primary sequence position (Protein Data Bank residue number). A chain identifier of a residue is not designated unless necessary. Bond lengths are in Å. Residues are underlined when the residue is in an α-helix, in italic when the residue is in a β-strand, or in ordinary font when the residue is in a coil location. Bold letters indicate that the residue side-chain atoms are buried (side-chain solvent accessibility is less than 10%). A residue in second shells is designated by the symbol ∗ when one of its side-chain atoms forms a hydrogen bond with a side-chain atom of a ligand; the symbol
when one of its main-chain atoms forms a hydrogen bond with a side-chain atom of a ligand; the symbol
when one of its side-chain atoms forms a hydrogen bond with a main-chain atom of ligand. Exclusive main chain and main chain hydrogen bonds are not indicated. Data set: a representative set of protein structures was based on the list of Hobohm and Sander (22), version of December, 1996, with pairwise primary sequence identity less than 25%. Total number of proteins in the list is 443 and those with metal, heme, or iron–sulfur linkage is 129 (29%). The structure data set was augmented with several recent protein structures known to contain metal centers.