Figure 1. Simplified structural diagrams for the YKO construct, pXP346, and pSO142-4.
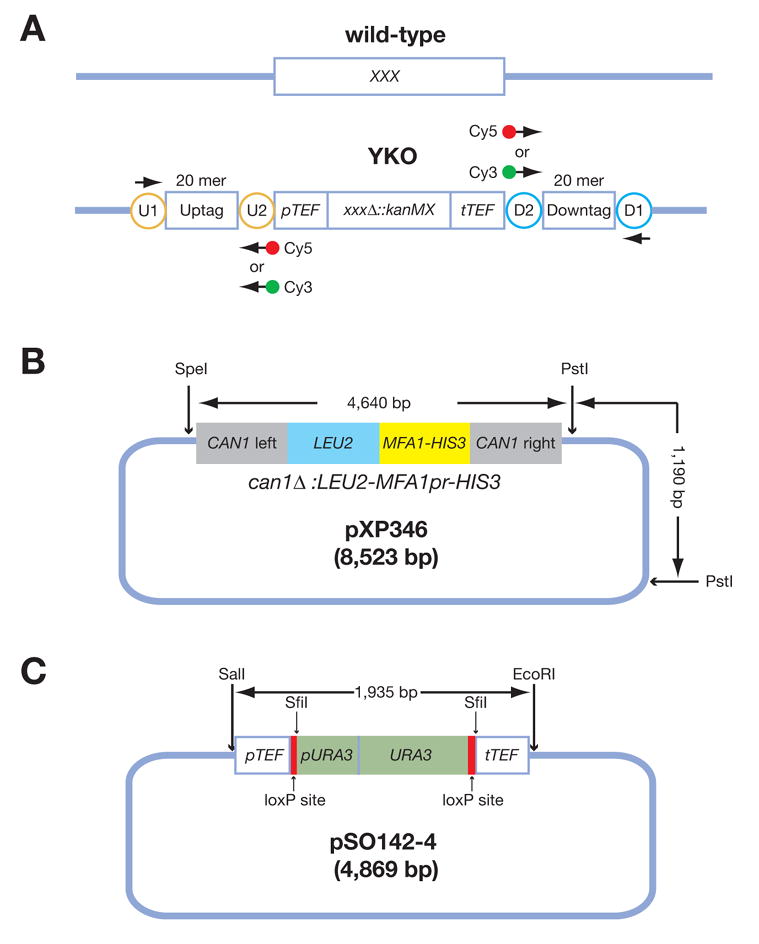
(A) A diagram for the yeast knockout construct. Each YKO consists of a kanMX module that confers resistance to the antibiotic G418 flanked by unique 20 mer “molecular barcodes” or “Tags” called the “Uptag” and “Downtag.” All “Uptags” are themselves flanked by a common set of priming sites (U1 and U2 within the orange circles) and all “Downtags” are flanked by another set of common priming sites (D1 and D2 within the cyan circles). These common priming sites allow for PCR amplification and microarray analysis of all Uptags or all Downtags in a population. “XXX” stands for any yeast gene. (B) A simplified diagram of pXP346, which contains the can1Δ::LEU2-MFA1pr-HIS3 reporter. This plasmid needs to be digested with SpeI and PstI to release the reporter for integration into the CAN1 locus of the yeast genome via homologous recombination. (C) A diagram of pSO142-4, which contains the URA3-loxP cassette. URA3 targeting construct pSO142 can be integrated into the existing kanMX4-marked YKO strains based on its sequence homology to the promoter and terminator sequences, derived from the Ashbya gossypii TEF gene. URA3, driven by its own promoter, is flanked by loxP sites allowing subsequent excision/marker swaps upon expression of Cre recombinase, increasing the flexibility of the marker for future manipulations. The SwaI and SfiI restriction sites were introduced for diagnostic purposes. In the first step, a loxP-URA3 cassette containing SwaI and SfiI sites was constructed using PCR. The loxP-URA3 cassette was amplified as two separate fragments, loxP-partial URA3 (loxP-URA3’) and partial URA3-loxP (‘URA3-loxP). The loxP, SwaI and SfiI sequences (introduced on primers) were fused to the 5′ and 3′ ends of the URA3 fragment via three sequential rounds of PCR. The URA3 fragment was amplified from pRS406. The loxP sequence was obtained from (25). Both PCR products were then co-transformed into a nej1 ::kanMX4 strain. The goal was to have the two PCR products homologously replace the kan ORF sequence. Integrative transformation was made possible by the following features on the two PCR products: loxP-URA3 contains 45 bp sequence homology to the TEF promoter, while URA3-loxP contains 48 bp sequence homology to the TEF terminator. The 3′ end of loxP-URA3’ and the 5′ end of ‘URA3-loxP overlap by 111 bp. nej1_ was used to decrease the efficiency of non-homologous end-joining. We first selected for Ura+ transformants. Next, we assayed and identified Ura+ transformants that are G418s, indicating that URA3 gene was integrated into kanMX4; one such strain was YSO205. In the second step, genomic DNA isolated from YSO205 was used as template to amplify the pTEF-loxP-URA3-loxP-tTEF fragment (1965 bp) using primers U2 and D2. The PCR products were then digested with SalI and EcoRI and cloned into pBSIIKS(−). Primer U2 contains a SalI site, while D2 contains EcoRI site.
Note: The plasmid backbones in both panels B and C are not drawn to scale.
