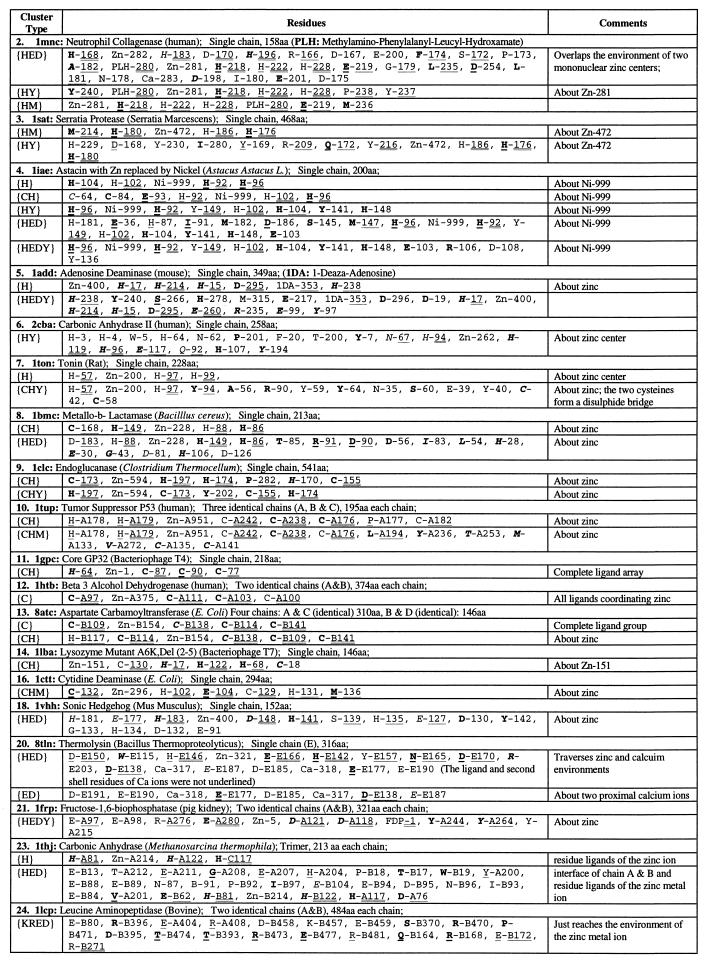
Table 2.
Significant residue clusters in protein 3D structures with single zinc ion center
The same sequential numbers of proteins are used as in Table 1. There is no statistically significant residue cluster (at 1% level) in atrolysin (1. 1atl), tramtrack zinc finger (15. 2drp), glycerol kinase (17. 1glc), carboxypeptidase (19. 2ctc), and enolase (22. 4enl). Procedures for identifying diverse residue clusters are described in the Methods sections of Karlin and Zhu (6) and Zhu and Karlin (7); see also the companion paper by Karlin et al. (1). A residue is described in two parts: part one, one-letter amino acid code and part two, a chain identifier (if any) and a residue number used in the Protein Data Bank file of the structure. The part one is underlined when the residue is in an α-helix; in italic when the residue is in a β-strand, and in ordinary font when the residue is in a coil. Boldface letters indicate that the residue side-chain atoms are buried (side-chain solvent accessibility less than 10%). Part two of a residue is doubly underlined when the residue is a ligand to the proximal metal ion or singly underlined when the residue is in the second shell of the proximal metal ion. Unless stated otherwise, a cluster is confined to one of identical chains and has analogs in the other chains.