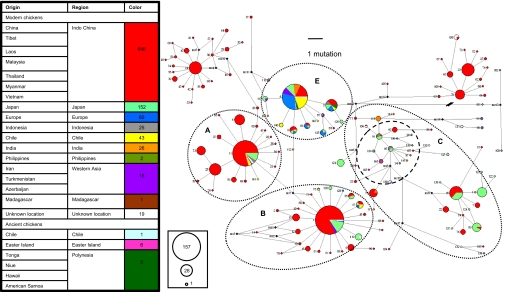
Fig. 1.
MJNs showing the relationships and clustering the mtDNA CR (205 bp) from worldwide, Chilean Araucana, pre-Columbian, and ancient Pacific/Polynesian chickens. Haplotype numbers are shown next to nodes, the geographical locations of samples are given in color, and node size is proportional to the frequency of the corresponding haplotypes as shown in the circles with numbers on the left. Branch lengths are proportional to the number of mutations except for two: the branch leading from median vector (mv) 12 to haplotype 101 (17 mutations) and the branch leading from mv 3 to haplotypes 13 and 19 (one mutation), for visual clarity. A, B, C, and E correspond to the major haplogroups identified by Liu et al. (6). Most of the modern and ancient Chilean chickens analyzed in this study cluster with Indian subcontinental/European/Chinese chickens (nodes 5, 8, 9, 128, and 140), with the remainder clustering with nodes predominant in South and eastern Chinese/Japanese/Indonesian chickens (nodes 11, 17, and 141). The pre-Columbian Chilean sequence falls within haplotype 8, which is the single most common chicken haplotype found around the world. Six ancient Polynesian sequences (Tonga Mele Havea and Ha'ateiho, Samoa Fatuma Futi, Hawaii Kualoa, Niue Paluki, and one from Easter Island) also cluster within the same haplogroup. Five of the ancient chicken sequences from Easter Island cluster with haplotypes 145 and 148 (dashed circle within haplogroup C), which are part of an uncommon group comprising mostly Indonesian chickens. The Easter Island sequences may represent mtDNA signatures of early Polynesian chicken transport.