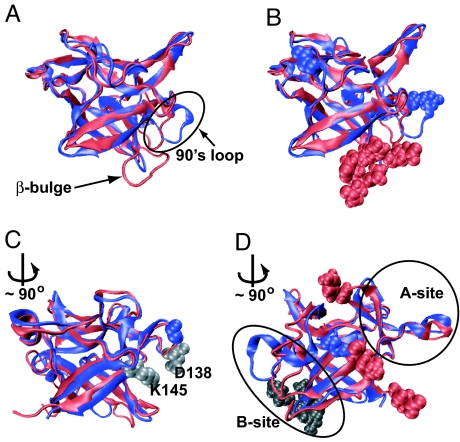
Fig. 2.
Different views of the structure alignment of the proteins IL-1β (red) and IL-1Ra (blue). Residues shown as red (IL-1β) and blue (IL-1Ra) spheres mark regions with the largest differences in structure. The rotation, when shown, is about the barrel axis and transforms the previous figure into the current one. (A) The two aligned proteins in approximately the same orientation as Fig. 1 A and B. The proteins are remarkably similar but an increased loop length is clearly visible in the β-bulge region. (B) The same orientation as in A. The structure of the loops in the 90s region is different even though the lengths of the loops are the same in the two proteins. The residue (G140) in the longer β11–β12 connecting loop of IL-1Ra is marked at the top left of the figure. (C) The gray spheres depict the interacting residues D138 and K145 that pin the connecting loop to the barrel. The loop is longer and has increased flexibility because of the insertion of G140 (shown as a blue sphere). (D) The three remaining structurally different residues are marked as red spheres. The cap residue is Y24. Residue 63 is on the cap/barrel interface and shows up as a change in structure in both proteins. In IL-1Ra, residue 63 interacts with the 90s loop. The terminal residue is S153. The β-bulge is shown (gray spheres) in the background for reference.