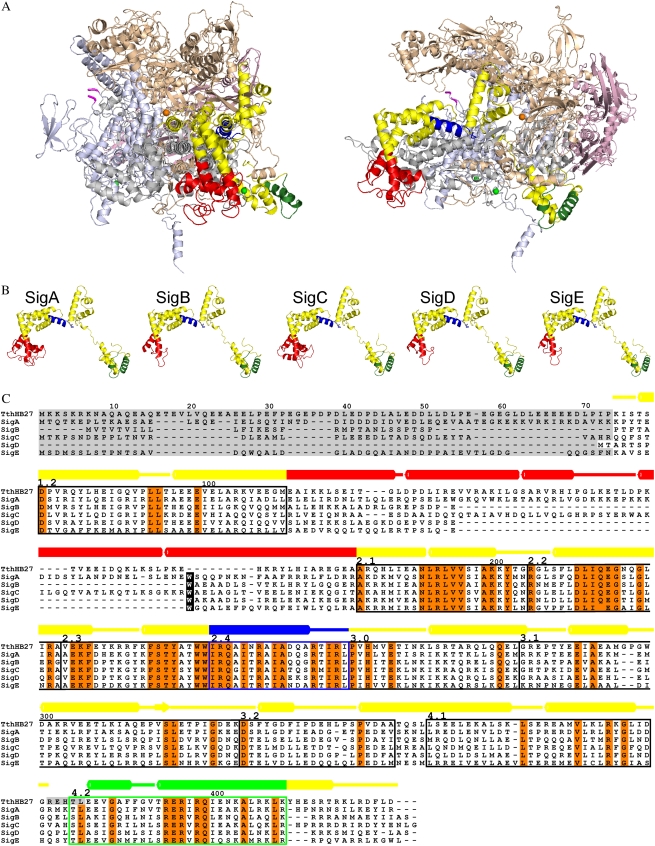
Figure 1.
Model of Synechocystis RNA polymerase. A, Homology modeling of the RNA polymerase holoenzyme with the SigA factor was performed using the crystal structure of the RNA polymerase holoenzyme of T. thermophilus (Artsimovitch et al., 2005; PDB code 2A6E; chains A–F) as the template. Views are from the front toward the catalytic center (left) and from the side (right). The core enzyme consists of two α subunits (light pink), the β subunit (light brown), the β′ subunit (light blue), and the γ subunit (gray). The first and last residues of the cyanobacterial insertion in the β′ subunit are indicated with magenta. In the otherwise yellow σ factor, the 4.2 region is green, the 2.4 region is blue, and the NCR connecting the conserved regions 1.2 and 2.1 is red. The catalytic magnesium ion is presented as an orange sphere, and two zinc ions are presented as green spheres. B, Models of principal (SigA) and group 2 (SigB–SigE) σ factors of Synechocystis. The coloring is as in A. C, Sequence alignment of Synechocystis and T. thermophilus σ factors. Absolutely conserved residues are shown in orange, and conserved regions from 1.2 to 4.2 are indicated with boxes. Residues missing from the structures are shaded gray. The numbering of amino acid residues and the secondary structure assignment, with barrels denoting α-helices, are shown above the alignment for the template. The coloring of secondary structures is as in A. The cyanobacterial conserved Trp residue is indicated with a black background.