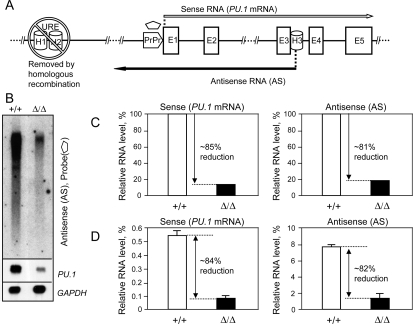
Figure 2.
The URE is instrumental in PU.1 antisense RNA synthesis. (A) The linear diagram shows the position of the PU.1 gene locus homology regions: the URE (H1 + H2), removed by homologous recombination (Rosenbauer et al. 2004), proximal promoter (PrPr), and H3; initiation sites and orientation of the sense (long white arrow) and antisense (long black arrow) transcripts; and the position of the antisense-specific probe (pentagon). (B) Northern blot analysis illustrates down-regulation of the antisense RNAs in the UREΔ/Δ mouse model (Rosenbauer et al. 2004). RNAs were isolated from bone marrow of wild-type (+/+) and hypomorphic (Δ/Δ) animals. Two micrograms of polyA(+) were run side by side and hybridized with the probe specific to antisense RNAs (pentagon, A). The top panel demonstrates down-regulation of antisense RNAs. The middle panel demonstrates down-regulation of the PU.1 sense mRNA in the UREΔ/Δ mouse model, using a double-stranded murine PU.1 cDNA probe. The bottom panel depicts GAPDH mRNA levels as loading controls (see the Supplemental Material for the probe information). (C,D) The URE regulates expression of both sense and antisense RNAs. Relative sense and antisense RNA levels in bone marrow of wild type and animals with an URE deletion were quantitated by Northern blot phosphorescence imaging (normalized to GAPDH mRNA) (C) and, as corroboration, by strand-specific RT–PCR (graph values calculated as mean ± standard deviation of three real-time RT–PCR runs, normalized to GAPDH mRNA, measured by TaqMan real-time analysis) (D). See the Supplemental Material for the TaqMan probes and primers used for strand-specific RT–PCR.