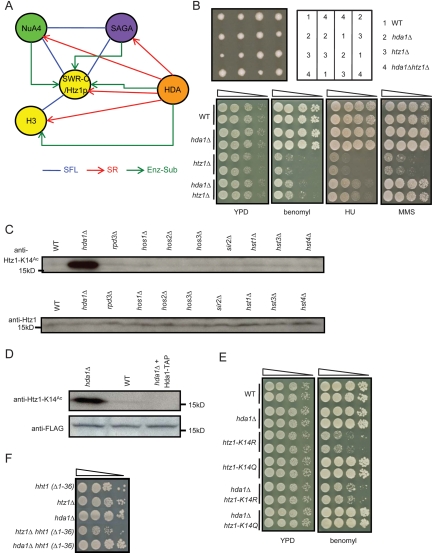
Figure 3.
HDA complex deacetylates Htz1-K14Ac in vitro and in vivo. (A) Overlap of the enzyme–substrate relationship with genetic interactions. A schematic model depicts genetic and enzyme–substrate interactions between HATs (NuA4 and SAGA), HDACs (HDA complex), and their substrates (Htz1p and histone H3). (B) Deletion of HDA1 rescues the slow-growth and drug-sensitivity phenotype of htz1Δ. Four tetrads dissected from an hda1Δ/HDA1 htz1Δ/HTZ1 diploid strain on YPD medium are shown. Drug sensitivities were assessed by plating 10-fold serial dilution on YPD medium containing HU (200 mM), MMS (0.03%), or benomyl (15 μg/mL). (C,D) Hda1p deacetylates Htz1-K14Ac in vivo and in vitro. The K14 acetylation level in wild-type and HDAC mutant strains was analyzed by immunoblot. (D) In vitro deacetylation activity of Hda1-TAP purified by tandem affinity purification was assessed by incubating it with Flag-tagged Htz1p purified from hda1Δ strain and then monitoring the K14 acetylation level by immunoblot. No protein was added to the control samples. (E) Deletion of HDA1 rescues the sensitivity of htz1-K14R to benomyl. The experimental conditions are described in B. (F) Genetic interactions of an N-tail deletion of histone H3 [hht1(Δ1–36)] with hda1Δ (alleviating) and htz1Δ (aggravating) mutants, suggesting that histone H3 reside in the same functional module as Htz1p.