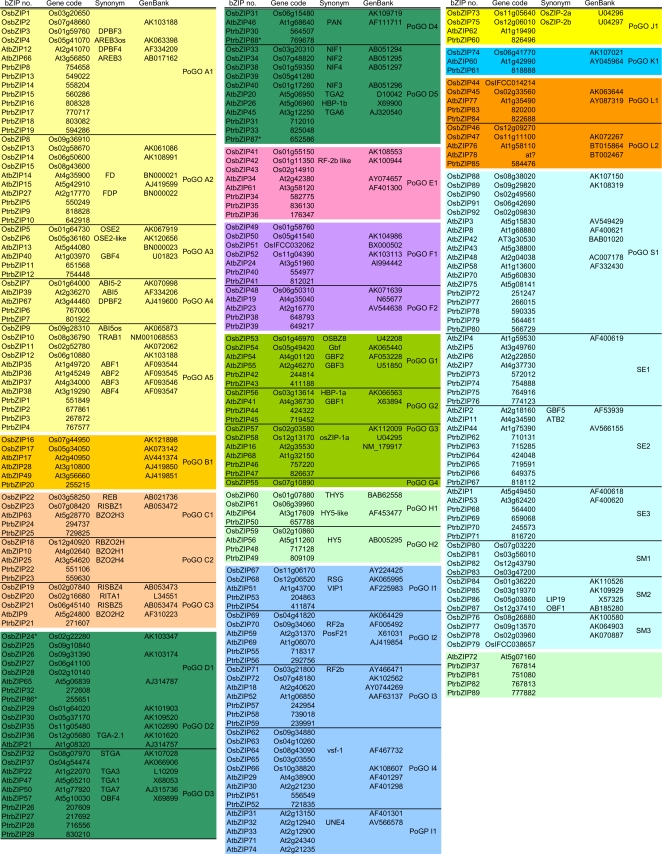
Figure 3. Classification of bZIPs from Arabidopsis, black cottonwood and rice.
Thirteen groups of homologues (A to L, and S) were defined through NJ phylogenetic analyses with the bZARP set (Figures S1 and S3). The organization into Possible Groups of Orthologues (PoGOs) was done by more refined NJ phylogenetic analyses inside each group of homologues, including also sequences from other eudicots and monocots. The alignment used for these analyses corresponds to a concatenated sequence of the group-specific conserved motifs identified employing MEME (http://meme.sdsc.edu/meme/website/intro.html; Figure 2). (*) Represents genes that lack group-wise conserved motifs, thus they were included inside a PoGO according to their best hit to another bZIP. Because the relation of AtbZIP72, PtrbZIP37, 81, 82 and 89 could not be clarified, they were not included in any of the groups of homologous or orthologous genes. One Possible Group of Paralogues (PoGP I1) was found in Arabidopsis. Column ‘Gene code’ provides the gene identifiers for Arabidopsis, black cottonwood and rice bZIP sequences taken from TAIR (http://www.arabidopsis.org/), JGI (http://www.jgi.doe.gov/) or TIGR (http://www.tigr.org/), respectively. ‘Synonym’ indicates published and often cites names of bZIP genes. The GenBank accession numbers of nucleotide sequences are given.