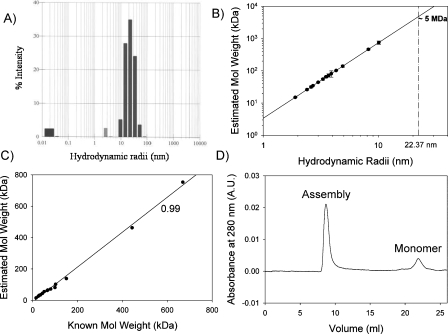
Figure 1.
(A) Histogram of distribution of hydrodynamic radii obtained from “regularization analysis” of data from dynamic light scattering experiments for 100 μM GED in 0.1 M phosphate buffer containing 1 mM EDTA, 150 mM NaCl at pH 5.7; average R h = 22.37 nm. (B) Plot of estimated molecular weight vs. hydrodynamic radii for 18 different proteins from literature (Claes et al. 1992); dashed line extrapolates the R h value of 22.37 nm on the fitted line (solid line) and corresponds to a molecular weight of ∼5 MDa. (C) Plot of estimated molecular weight vs. known molecular weight for the 18 proteins used in plot B; correlation coefficient between the two is found to be 0.99. (D) Size-exclusion chromatogram of GED (100 μM, 100 μL) in 0.1 M phosphate buffer containing 1 mM EDTA, 150 mM NaCl at pH 5.7, run on a Superose 6 10/300 GL column (GE Healthcare) using the BioLogic FPLC system (Bio-Rad), at a flow rate of 0.5 mL/min.