Figure 1.
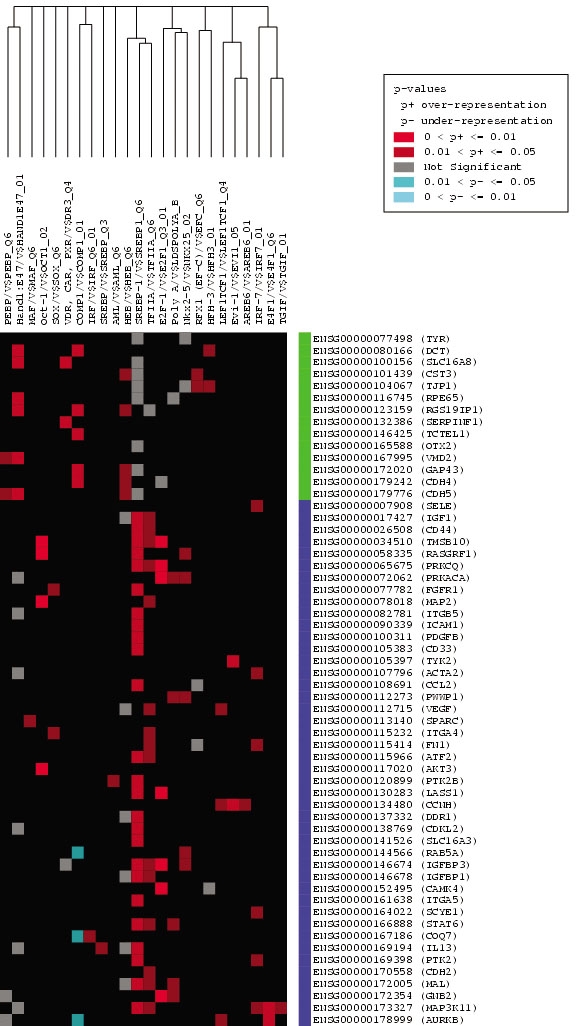
Candidate interaction matrix for statistically enriched transcription response elements from promoter analysis and interaction network toolset analysis of human gene promoters. The retinal pigment epithelium (RPE) gene set was analyzed by promoter analysis and interaction network toolset (PAINT) and a graphic candidate interaction matrix (CIM) was generated as described in Methods. The y-axis lists the Ensembl Gene identifiers for each gene and the x-axis lists the TRANSFAC identifiers for each transcription response element (TRE) found at least once in the promoter region of one or more genes. Genes listed along the y-axis are divided into two clusters that are either upregulated (blue) or down-regulated (green) during epithelial-mesenchymal transformation (EMT) of RPE cells. TREs listed along the x-axis are clustered according to related occurrence pattern calculated using Jaccard's coefficient. The elements within the matrix are color-coded based on the p-value of each TRE found in the regulatory regions of the genes. A red dot represents a TRE that is statistically significant and therefore over-represented in our gene set, while a blue dot signifies an under-represented TRE and a gray dot stands for a TRE with no statistical significance in our gene list. This figure represents the subset of enriched TREs for the human genome; the full CIMs for human and other genomes analyzed are shown in Appendix 4, Appendix 5, Appendix 12, and Appendix13.
