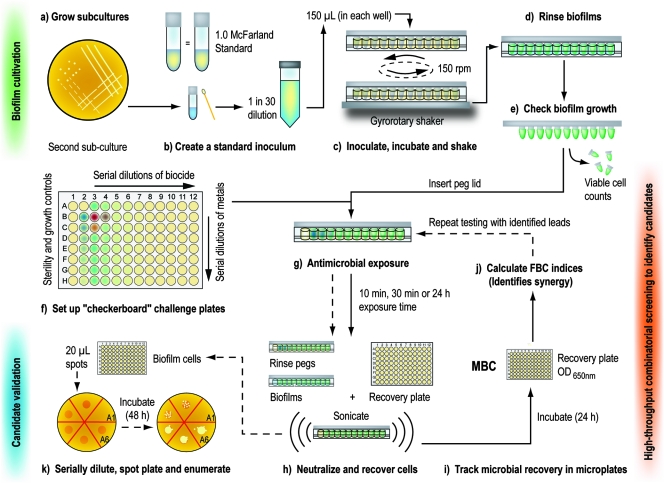
FIG. 1.
High-throughput screening may be used to identify synergistic antimicrobial interactions that kill microbial biofilms. Starting from cryogenic stocks, the desired bacterial strain was streaked out twice on TSA (a), and colonies from the second subcultures were suspended in growth medium to match a 1.0 McFarland optical standard (b). This standardized suspension, diluted 30-fold in TSB, served as the inoculum for the CBDs. The inoculated devices were assembled and incubated on a gyrorotary shaker (c), which facilitated the formation of 96 statistically equivalent biofilms on the peg surfaces (data not shown). Biofilms were rinsed with 0.9% NaCl (d), and surface-adherent growth was verified by viable cell counting (e). Antimicrobials were set up in “checkerboard” arrangements in microtiter plates (f), and the rinsed biofilms were inserted into these challenge plates for the desired exposure time (g). Following antimicrobial exposure, biofilms were rinsed and inserted into recovery plates. Biofilm cells were disrupted into the recovery medium by sonication (h), and the recovery plates were incubated for 24 h before the OD650 values of recovered cultures were read in a microtiter plate reader (i). This allowed the FBC index to be calculated, and this was used to identify “candidate” synergistic interactions (j). Candidates were validated by repeating the testing process (as outlined in steps a to h), but instead of qualitative measurements, biofilm cell survival was quantified by viable cell counting on agar plates (k).