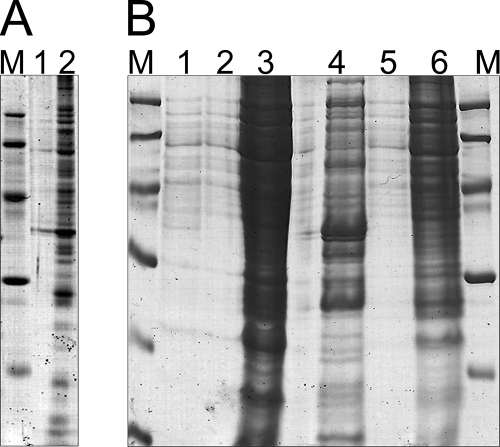
FIG. 1.
Optimization of the protein extraction protocol. (A) Protein extraction was performed by classical treatment with phenol/acetone starting from sedimented cells previously cultivated in MM under conditions promoting PHB accumulation, and it showed disturbance of protein extraction by PHB. Protein pellets were dissolved in rehydration buffer A, and for both samples, the polyacrylamide gel (12% [wt/vol]) was loaded with 15 μl protein solution and 5 μl denaturation buffer (0.225 M Tris-HCl [pH 6.8], 50% [wt/vol] glycerol, 5% [wt/vol] SDS, 0.05% [wt/vol] bromophenol blue, 0.25 mM DTT). Lane 1, R. eutropha H16; lane 2, R. eutropha PHB−4. (B) Application of the modified protein extraction protocol, including ultracentrifugation, which was developed in this study. Lane 1, R. eutropha H16, supernatant after ultracentrifugation (for details, see “Preparation of protein samples for proteome analysis” in the text); lane 2, R. eutropha H16, sedimented proteins from ultracentrifugation after resuspension in crack solution; lane 3, R. eutropha H16 after protein extraction from the supernatant and resuspension of the resulting protein pellet in crack solution; lane 4, R. eutropha PHB−4, supernatant after ultracentrifugation; lane 5, R. eutropha PHB−4, sedimented proteins from ultracentrifugation after resuspension in crack solution; lane 6, R. eutropha PHB−4, after protein extraction from supernatant and resuspension of the resulting protein pellet in crack solution. For each lane, 15 μl of the solution was loaded, together with 5 μl denaturation buffer. Lanes M, molecular mass markers.