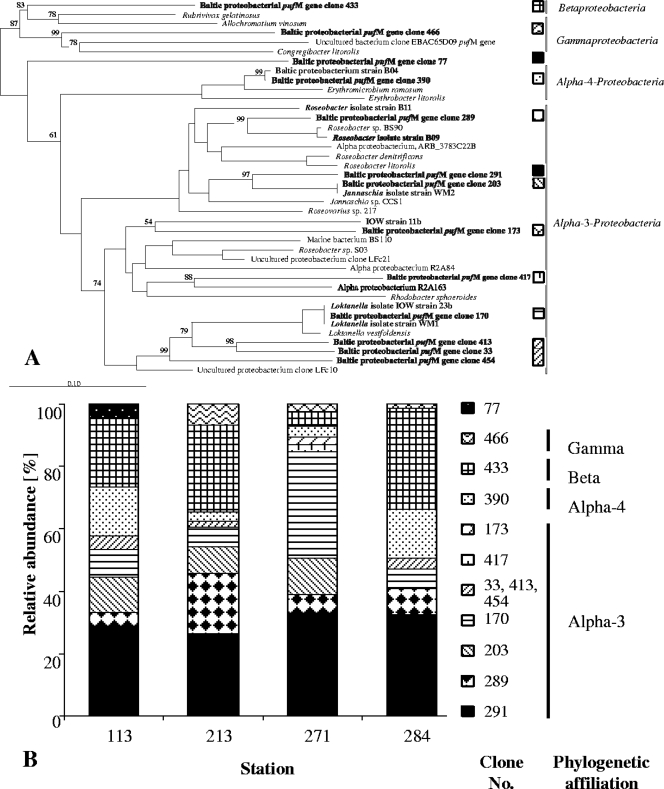
FIG. 3.
(A) Unrooted tree showing the phylogenetic relationships of pufM gene sequences detected in the central Baltic Sea and their closest relatives. The tree was reconstructed by using the neighbor-joining method and is based on a comparison of approximately 400 nucleotides. Bootstrap values, expressed as a percentage of 500 replications, are given at branching points. Only values greater than 50% are shown. Chloroflexus aurantiacus was used as the outgroup. The pufM sequence of L. vestfoldensis DSM16212T was obtained in the present study. Names in boldface indicate clones or isolates obtained from the present study. Patterns indicate representative clones that were retrieved throughout the stations. Bar, 10 substitutions per 100 nucleotides. (B) Relative abundance of different pufM genes in clone libraries, reflected by patterns identical to those from panel A. Clones derived from the depths of the maximal AAP concentration of the Arkona Basin (station 113), Bornholm Basin (station 213), Gotland Basin (station 271), and Landsort Deep (station 284).