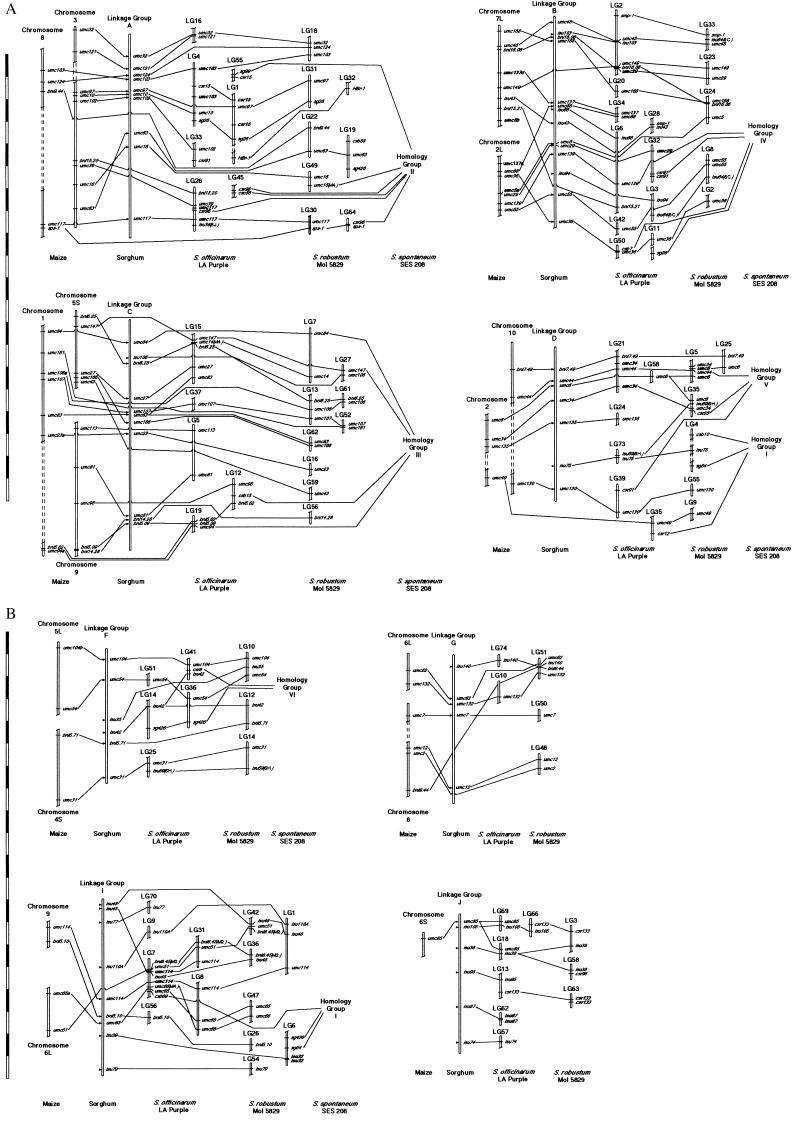
Figure 1.
Comparative genetic maps of maize, sorghum, and three species of Saccharum. DNA marker positions in maize were based on the UMC maize RFLP map (http://teosinte.agron.missouri.edu). Sorghum linkage groups (LGs) were named according to Chittendenn et al. (44). Sorghum marker positions were compiled based on Whitkus et al. (12), Pereira et al. (43), Lin et al. (20), and Paterson et al. (21); their map locations are indicated by an arrow. S. spontaneum ‘SES 208’ homeology groups (HGs) were based on daSilva et al. (5). DNA markers whose positions conflict with the sorghum comparisons have their location indicated in parenthesis, using “S” for sorghum and “M” for maize. Linkage groups were formed by using RFLP, AFLP, and arbitrarily primed PCR markers, but those shown are only the RFLP markers because they enabled the comparisons. Probes that have been mapped in a species have their position indicated with a line that traverses the chromosome or linkage group, whereas probes whose positions are unknown (i.e., not mapped) contain lines that pass behind the chromosome or linkage group. Maize probes unmapped in sorghum have arrows indicating their inferred position in sorghum (based on combining information from previous maps). Breaks in the LGs or chromosomes indicate discontinuity of the LGs or chromosomes. For maize chromosomes, S = short arm and L = long arm. (Bar = 10 cM.)