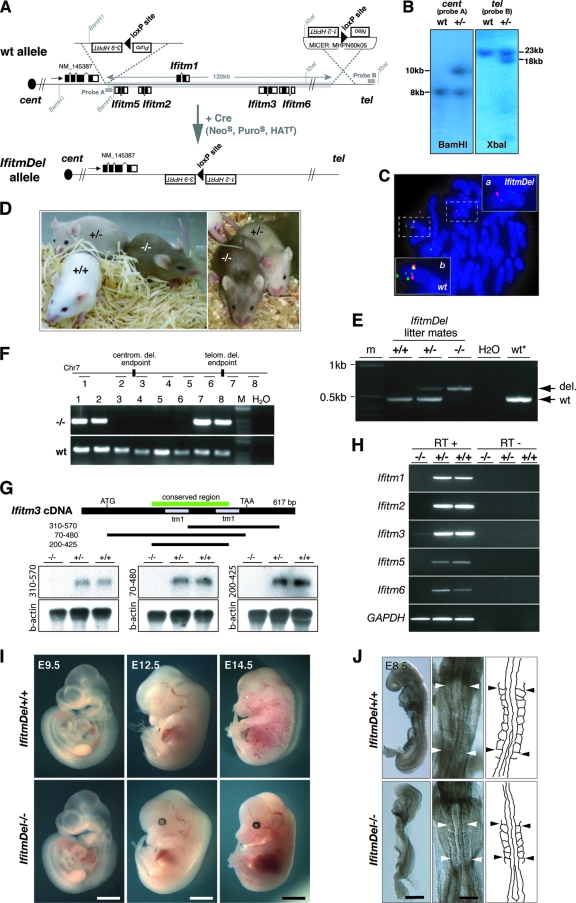
FIG. 1.
Targeted deletion of the mouse Ifitm gene cluster. (A) Schematic of the targeting strategy used to create the IfitmDel allele. (B) Southern blot analysis of targeted ES cell clones prior to Cre-mediated excision. (C) FISH analysis of IfitmDel+/− ES cell metaphase spreads using a fluorescein isothiocyanate-labeled genomic probe in the deletion (green) and a Texas red-labeled BAC-derived probe outside the deletion (red). Inset a, IfitmDel allele; inset b, wt allele. (D) Fourteen-day-old IfitmDel−/−, IfitmDel+/−, and wt littermates. Coat color marking of the targeted allele enables genotyping by eye. (E) Confirmation of genotyping by PCR on tail extracts. (F) PCR on genomic DNA of IfitmDel−/− and wt tissues using primers amplifying fragments flanking (lanes 1, 2, 7, and 8) and within (lanes 3 to 6) the deletion confirms deletion of the Ifitm locus from the targeted genome. (G) Northern blot analysis using E17.5 whole-embryo RNA from IfitmDel−/−, IfitmDel+/−, and wt littermates. Three probes corresponding to different fragments of Ifitm3 cDNA were used for hybridization: 310 bp to 570 bp, Ifitm3-specific probe; 70 bp to 480 bp, Ifitm3 coding region probe, expected to cross-hybridize with all known mouse Ifitm transcripts; 200 bp to 425 bp, Ifitm gene highly conserved region probe, expected to cross-hybridize with any Ifitm-like transcript. (H) Reverse transcription-PCR analysis on E17.5 whole embryo cDNA from IfitmDel−/−, IfitmDel+/−, and wt littermates using Ifitm gene-specific primer sets. (I) IfitmDel−/− and wt littermates at different stages of development. (J) E8.5 IfitmDel−/− embryos show no defects in somite organization. Left panels, lateral view; middle panels, dorsal view; right panels, outline of somites and neural tube in dorsal view. Bars, 400 μm (I, left panels), 2.25 mm (I, middle panels), 3.25 mm (I, right panels), 200 μm (J, left panels), and 100 μm (J, middle panels).