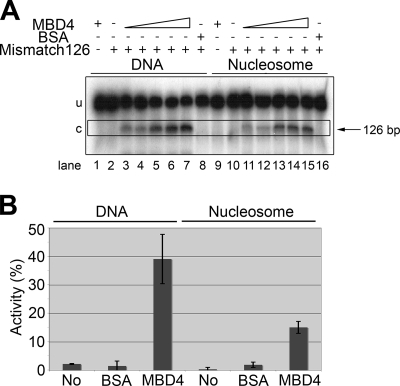
FIG. 7.
MBD4 glycosylase activity in the presence of a nucleosome. (A) Urea-denaturing PAGE analysis of the glycosylase activities on a 164-bp DNA construct with a mismatch at position 126 in its DNA and reconstituted nucleosome versions. Approximately 10 nM DNA was incubated with MBD4 at 10 nM (lanes 3 and 11), 20 nM (lanes 4 and 12), 40 nM (lanes 5 and 13), 80 nM (lanes 6 and 14), and 160 nM (lanes 1, 7, 9, and 15) or with 160 nM BSA (lanes 8 and 16) (see Materials and Methods for further details). Because the nucleosome reconstitution procedure was carried out in the presence of a 10-fold excess of a 146-bp random-sequence DNA fragment (see Materials and Methods), the assay of glycosylase activity on naked DNA was also carried out in the presence of a 10-fold excess of the same 146-bp random-sequence cold DNA. Triangles indicate increasing concentrations. u, uncut DNA substrate; c, cut DNA product; −, absent; +, present. (B) Bar plot representation of the glycosylase activity indicated in panel A for 160 nM MBD4. The activity is expressed in terms of the amount of the NaOH-cut 126-bp fragment as a percentage of the amount of the total starting mismatched DNA template (cut plus uncut). “No” indicates a 164-bp DNA template with no mismatch.