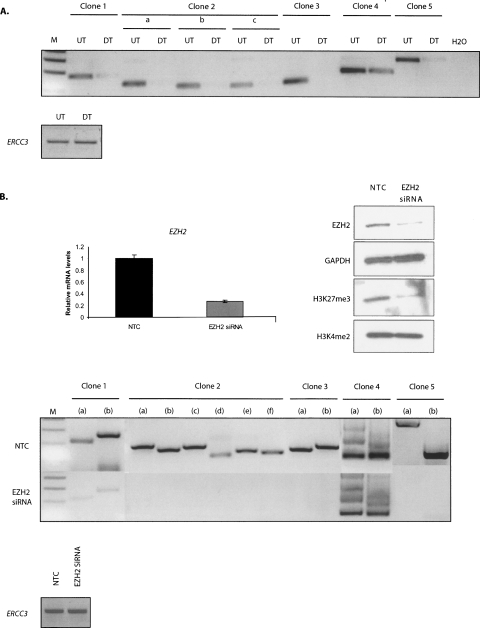
Figure 2.
EZH2 mediates long-range inter- and intrachromosomal interactions in undifferentiated Tera-2 cells. (A) 6C interactions exist in vivo in undifferentiated Tera-2 cells and are lost on ATRA-induced cellular differentiation toward neuronal lineage. Semiquantitative 3C PCRs were performed using one primer from each partner in the clone to detect interactions between 6C partners in undifferentiated Tera-2 (UT) and differentiated Tera-2 (DT) cells. For clone 2, “a” refers to the interaction between partner 1 and partner 2; “b,” the interaction between partner 1 and partner 3; and “c,” the interaction between partner 2 and partner 3. In the lower panel; interaction between two fragments at the ERCC3 locus, separated by ∼10 kb, was used to normalize crosslinking frequency between UT and DT samples. (B) siRNA-mediated depletion of EZH2 leads to loss of 6C interactions. The upper panel shows the elimination of the target mRNA (left panel) and protein (right panel) by siRNA against EZH2. As a control, a random-sequence nontargeting control siRNA was used (NTC). The undifferentiated Tera-2 cells were transfected with 25 nM EZH2 siRNA for two consecutive days using Lipo 2000 reagent and were processed 72 h later for real-time RT-PCR analysis using EZH2 and GAPDH-specific primers (as controls) (upper left panel) and for Western analysis using anti-EZH2 and anti-H3K27me3 antibodies (upper right panel). The blot was subsequently stripped and reprobed with anti-GAPDH and H3K4me2 antibodies as controls. The H3K4me2 marks remain unaffected, indicating that the targeting is specific for EZH2, which catalyzes the H3K27me3 marks. 3C assay was carried out in undifferentiated Tera-2 cells 72 h post-transfection with either siRNA specific for EZH2 or with a NTC (lower panel). Semiquantitative 3C PCRs were performed using multiple primer combinations to detect interactions between 6C partners. For the 3C PCRs, following combinations of primers (FP indicates forward primer; RP, reverse primer) were used. For clone 1, (a) FP of partner 1 and FP of partner 2, (b) RP of partner 1 and RP of partner 2; for clone 2, (a) FP of partner 1 and FP of partner 2, (b) FP of partner 1 and FP of partner 3, (c) RP of partner 1 and RP of partner 2, (d) RP of partner 1 and RP of partner 3, (e) FP of partner 2 and FP of partner 3, (f) RP of partner 2 and RP of partner 3; for clone 3, (a) FP of partner 1 and FP of partner 2, (b) RP of partner 1 and RP of partner 2; for clone 4, (a) FP of partner 1 and FP of partner 2, (b) RP of partner 1 and RP of partner 2; and for clone 5, (a) FP of partner 1 and FP of partner 2, (b) RP of partner 1 and RP of partner 2. In the lower panel; interaction between two fragments at ERCC3 locus, separated by ∼10 kb, was used to normalize crosslinking frequency between NTC and EZH2 siRNA-treated samples. The figure represents the results of one experiment, which were further confirmed with samples generated in independent experiments.