Figure 1.
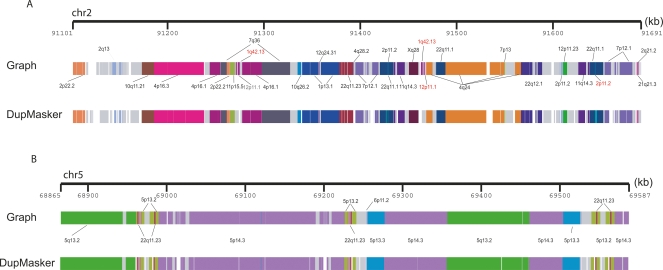
DupMasker defines the substructure of human segmental duplication blocks. Human segmental duplications are organized into complex duplication blocks where individual duplicons originate from different regions of the genome. We assessed the ability of DupMasker to accurately define these ancestral duplicons in this context by comparing results from two regions studied previously in detail (Horvath et al. 2000; Jiang et al. 2007). We schematically display (PARASIGHT: http://eichlerlab.gs.washington.edu/jeff/parasight/index.html) duplicons detected using the A-Bruijn graph approach (top) versus DupMasker (bottom) for (A), an ∼600-kbp region on chromosome 2p11 and (B), an ∼700-kbp region on chromosome 5q13.2. The different duplicons are illustrated as color-coded blocks; different colors correspond to different cytogenetic band locations of the ancestral loci. We found 33/36 nonredundant duplicons blocks are consistent between these two results. The three mismatched blocks are relatively small in length (length <1.5 kb, highlighted in red).