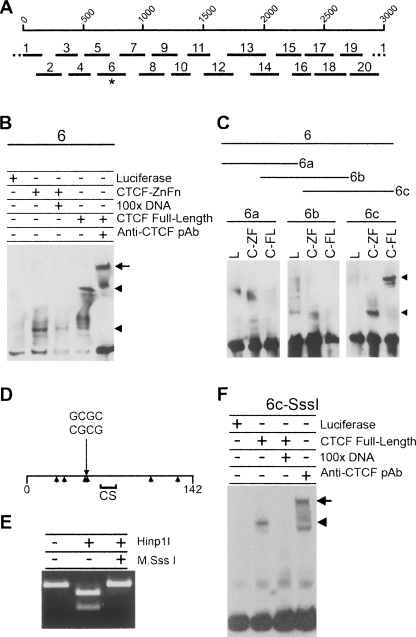
Figure 5.
CTCF binds to DXZ4 DNA in vitro independent of flanking CpG methylation. (A) A single 3-kb DXZ4 monomer is represented by 1–3000 bp scale, under which are shown the 20 different fragments used in the original EMSA analysis (see Supplemental Fig. 2). The asterisk indicates fragment-6, the mobility of which is retarded in the presence of CTCF protein. (B) Confirmation of CTCF specificity by 100× excess competition with unlabeled 6-DNA and supershift (upper arrowhead and arrow, respectively). (C) Refining CTCF to 6c only in the presence of the CTCF zinc fingers region (C-ZF) and full-length CTCF (C-FL) as indicated by the arrowheads, but not by luciferase (L). (D) Map of fragment 6c. The 142-bp fragment 6c is represented by the horizontal line, with the location of the Hinp1I site (GCGC) indicated. Arrowheads below the line indicate location of CpG sequences. The region denoted CS indicates a region with a good match to the CTCF genome-wide consensus sequence (Kim et al. 2007). (E) In vitro methylation of fragment 6c by M.SssI CpG methyltransferase blocks digestion by the methyl-sensitive Hinp1I restriction endonuclease. (F) CTCF binding to CpG methylated fragment 6c (Me-6c).