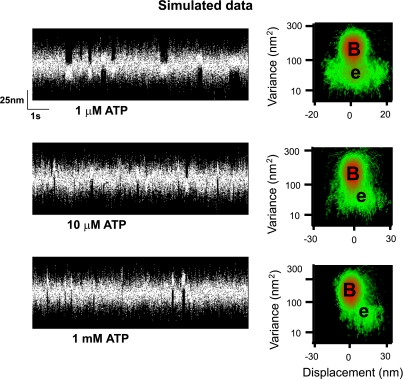
Fig. 4.
Simulated single molecule data. Single molecule data were simulated with two kinds of binding events: one that binds actin productively and generates a full displacement (10 nm) and a second, rigor-like population, that binds actin but generates no net displacement (0 nm), having a very broad displacement distribution. The simulations were performed using custom software previously described (12) with the ratio of nonproductive to productive events set at 6 to 1. In the simulations the ADP release rate was set to 33.s−1 (Fig. 3), whereas the rigor state lifetime was varied to simulate increasing ATP concentrations based on a second-order ATP-dependent dissociation constant of 5 × 106 .M−1·s−1 (Fig. 3). The lifetime of the productive binding events was the sum of the ADP and rigor state lifetimes, whereas the nonproductive events were determined by only the rigor lifetime. At 1 μM ATP, the event population is broadly distributed (see MV histogram) giving a value for d (2.5 nm) equivalent to that observed at pH 6.4 (and 1 μM ATP) for real data (see Fig. 2). However, with increasing ATP concentrations, the lifetime of the nonproductive events become shorter and barely evident at 10 μM ATP. The step size for the 10 μM ATP condition is 6 nm, similar to the real data (see Table 1). At 1 mM ATP, the nonproductive events are below the detection limits of the MV analysis so that only the productive, 10-nm population remain, and thus restore the step size back to its 10-nm normal value.