Figure 3.
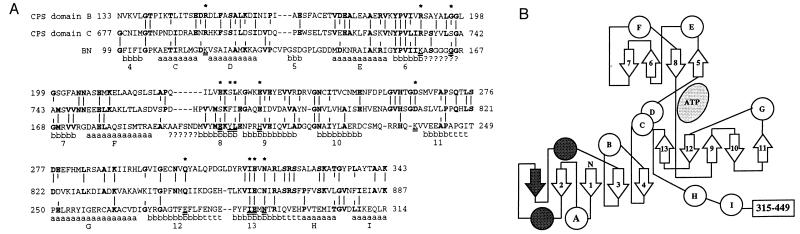
(A) Alignment of yeast arginine-specific CPS domain B (residues 133–343) and domain C (residues 677–887) with BN (residues 99–314; ref. 42). The alignment is essentially that of Simmer et al. (2) and is based on alignment of five different CPSs. Vertical lines indicate sequence identity between the CPS domains or between BN and one or both of the CPS domains; the identical residues are shown in bold. a, b, and t indicate α-helix, β-sheet, and turn structural elements, respectively, in the solved structure of BN, and ? indicates residues too disordered to be included in the solved structure (24). The 12 BN residues conserved or conservatively substituted in the ATP binding sites of BN, GS, and AA (26) are indicated by underlining of the BN residues and by stars above their positions. (B) Topological diagram of residues 1–314 of the BN structure, simplified from Artymiuk et al. (26). α-Helix elements are shown as circles and β-sheet elements as arrows. The unshaded elements (helices A–I and sheets 1–13; also indicated on the bottom line of A) are those that are found in all members of the N-ligase family and the shaded elements are those unique to BN (26). The topology of residues 315–449 (data not shown) is also unique to BN.