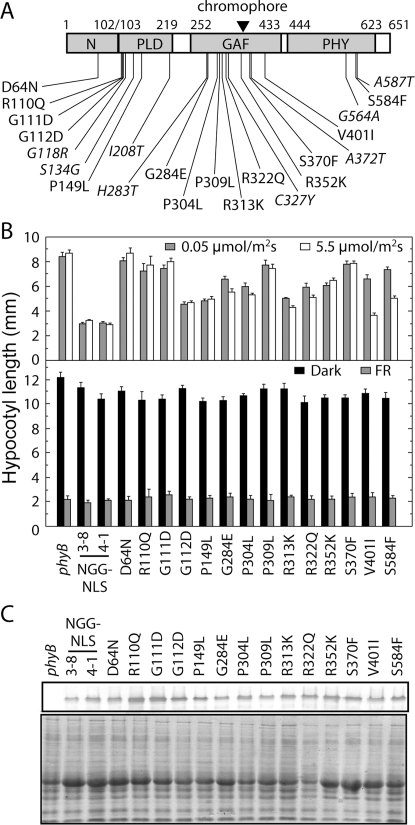
Figure 1. Hypocotyl Phenotypes of N651-GUS-NLS Mutants Carrying Missense Mutations.
(A) Locations of missense mutations found in the present (plain) and previous (italic) studies. For details, see Table 1. PLD, GAF and PHY were delimited as amino acid residues 103–219, 252–433 and 444–623, respectively, according to a sequence-based domain database, Pfam version 20.0 (http://www.sanger.ac.uk/Software/Pfam). The N-terminal extension was defined as amino acid residues 1–102. The closed triangle represents the chromophore binding site. (B) Hypocotyl lengths of mutants grown under different light conditions. For the hypocotyl measurement, plants were grown under weak cR (0.05 µmol m−2 sec−1) (shaded, upper panel), strong cR (5.5 µmol m−2 sec−1) (open, upper panel), cFR (10 µmol m−2 sec−1) (shaded, lower panel) or in darkness (closed, lower panel). The mean±SE (n = 25) is shown. (C) Immunoblot detection of the N651G-GUS-NLS proteins. For detection, 50 µg of total protein was loaded in each lane, blotted onto nitrocellulose membrane after SDS-PAGE, and probed with an anti-GFP monoclonal antibody (SIGMA) (upper panel). To confirm protein loading amount, the same samples were subjected to Coomassie Brilliant Blue (CBB) staining (lower panel).