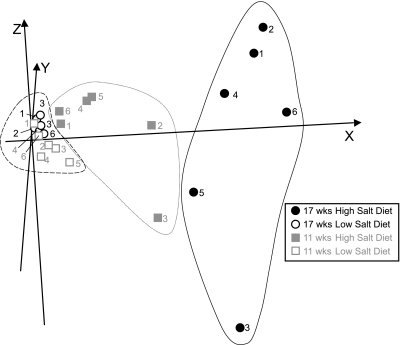
Fig. 3.
Segregation of animals with cardiac dysfunction from controls using 30 selected genes. The 20 out of 30 genes in Table 3 with expression were correlated with FS by regression analysis and were used for principal component analysis (PCA). PCA is used to present animals with similar gene expression profiles close in three-dimensional space. Because PCA is performed based on anonymously transformed gene expression data from each animal, each axis has only mathematical relevance for each spot (animal). Seventeen-week-old HT (area in far right black line) and NT (area in far left dotted line) animals were successfully segregated, and the 11-wk-old HT animals (area in middle gray line) were segregated to a third group between these 2 above groups. Individual animals are identified by numbers, as in Fig. 1.