Abstract
Uncertainty as to which member of a family of DNA-binding transcription factors regulates a specific promoter in intact cells is a problem common to many investigators. Determining target gene specificity requires both an analysis of protein binding to the endogenous promoter as well as a characterization of the functional consequences of transcription factor binding. By using a formaldehyde crosslinking procedure and Gal4 fusion proteins, we have analyzed the timing and functional consequences of binding of Myc and upstream stimulatory factor (USF)1 to endogenous cellular genes. We demonstrate that the endogenous cad promoter can be immunoprecipitated with antibodies against Myc and USF1. We further demonstrate that although both Myc and USF1 can bind to cad, the cad promoter can respond only to the Myc transactivation domain. We also show that the amount of Myc bound to the cad promoter fluctuates in a growth-dependent manner. Thus, our data analyzing both DNA binding and promoter activity in intact cells suggest that cad is a Myc target gene. In addition, we show that Myc binding can occur at many sites in vivo but that the position of the binding site determines the functional consequences of this binding. Our data indicate that a post-DNA-binding mechanism determines Myc target gene specificity. Importantly, we have demonstrated the feasibility of analyzing the binding of site-specific transcription factors in vivo to single copy mammalian genes.
The study of mammalian gene transcription is complicated by the fact that many regulatory sequences are recognized by multiple members of the same transcription factor family. For example, in vitro-binding assays have demonstrated that the E box motif (CACGTG) is the binding site for members of the basic helix–loop–helix leucine zipper (bHLHzip) family. The expression of many genes has been shown to be influenced by the presence of one or more E box elements. Proteins that bind to E boxes include members of the Myc network such as the heterodimeric complexes Myc/Max (1), Mad/Max (2), Mxi/Max (3), and Mnt/Max (4), as well as other bHLHzip proteins such as USF (5), TFE3 (6), and TFEB (7). Several E box-containing genes have been classified as targets of Myc such as odc (8, 9), α-prothymosin (10), ECA39 (11), eIF4E (12), cdc25 (13), MrDb (14), and cad (15, 16). However, it remains unclear whether Myc actually regulates these genes in vivo, under physiologic conditions.
The lack of in vitro-binding specificity between members of the bHLHzip family might be interpreted as evidence that E box-binding transcription factors are functionally redundant. However, in vivo studies do not support this conclusion. The observation that Myc, but not USF1, cooperates with Ras to transform cells supports a model in which discrimination between E box-binding family members does occur at certain genes in vivo (17). Although little is known about how target gene specificity is achieved, two basic hypotheses exist. First, it is possible that DNA-binding specificity between family members occurs at certain E box elements in vivo. Second, a post-DNA-binding mechanism may regulate target gene activation. In support of the first model, the context of the E box hexanucleotide with respect to immediate flanking sequences can determine DNA-binding specificity between certain bHLHzip proteins (18). Alternatively, as evidence for a post-DNA-binding mechanism, the position of the E box with respect to the transcriptional start site has been suggested to be important for accurate selection and regulation of gene expression (10). However, none of the previous studies directly examined which E box-binding protein binds to and regulates transcription from a specific E box element in vivo.
To address the question of target gene specificity, we required a well-characterized model system. Previously, we demonstrated that transcription from the hamster cad (carbamoyl-phosphate synthase/aspartate carbamoyltransferase/dihydroorotase) promoter is growth responsive. The growth regulatory element was mapped to a consensus E box located 65 bp downstream of the transcriptional start site (15). Expression from the cad promoter during the growth cycle appears to require Myc because coexpression of Myc dominant-negative proteins reduce cad growth regulation (15). However, these studies did not directly analyze protein binding to the cad promoter during the growth cycle.
Here, we have used a modified formaldehyde crosslinking procedure to evaluate growth phase-specific binding of Myc and USF1 to the cad promoter in mouse NIH 3T3 cells. The successful adaptation of this procedure has allowed us to investigate whether target gene specificity exists and whether the key step in Myc-mediated transactivation occurs at DNA binding or at a post-DNA-binding step. Our results show that the amount of Myc bound to the cad promoter increases after serum stimulation of quiescent cells whereas USF1 binding is constitutive. Because both Myc and USF1 bind to the cad promoter in S phase cells when the promoter is active, we propose that a post-DNA-binding mechanism determines target gene specificity. In support of this model, we show that the transactivation domains of Myc and USF1 are not equivalent in their ability to activate cad expression. Additionally, we show that Myc binds to many sites in vivo but that the position of the binding site can determine the functional consequences of this binding.
MATERIALS AND METHODS
RNase Protection Assay.
RNase protection assays and preparation of RNA were carried out as described (19). A 209-nt 32P-labeled RNA probe was transcribed with T7 RNA polymerase from a HindIII-linearized pMcad plasmid. pMcad contains a 154-bp fragment of the mouse cad cDNA, which was cloned from NIH 3T3 RNA by reverse transcriptase–PCR. The cloned murine cad cDNA fragment was submitted to GenBank (accession no. AF053339). Cytoplasmic RNA (10 μg) prepared from serum-starved or serum-stimulated NIH 3T3 cells was hybridized with 105 cpm probe at 65°C for 3 hr. Unhybridized RNA was digested by the addition of 10 μg of RNase A. The products were resolved on a 6% denaturing polyacrylamide gel and visualized by autoradiography.
Cell Culture and Transfections.
NIH 3T3 cells were maintained as described (16). Serum synchronization transfections were performed as described (16). For cad-reporter cotransfections, 1.5 × 105 cells were transfected with 0.5 μg of the cad Gal4 site-containing reporter, 2.5 μg of Gal4 fusion protein expression plasmid, and sheared salmon sperm DNA to a total of 15 μg of DNA. Identical reactions were prepared for the dihydrofolate reductase (dhfr) cotransfections except that 2.5 μg of the dhfr Gal4 site-containing reporter and 5.0 μg of the plasmid which expresses the Gal4 fusion protein were used. After transfection, cells were maintained in growth media (DMEM + 5% bovine calf serum) and harvested after 48 hr.
Plasmids.
The plasmid pMcad2 contains a 659-bp fragment of the mouse cad promoter that was amplified from C57BL/6J mouse genomic DNA by using PCR primers against regions of highly conserved sequence between the hamster (20) and human (S. Mac and P.J.F., unpublished results) cad promoters cloned into SmaI-digested pBSM13+. The pMcad2 was sequenced by using AmpliTaq DNA polymerase FS, Dye-Terminator chemistry (Perkin–Elmer), analyzed by using abi prism sequencing software (Applied Biosystems), and submitted to GenBank (accession no. AF053338). The mouse cad reporter plasmids (mcad) −148/+84luc and mcad −148/+57luc were cloned from the pMcad2 plasmid by PCR using a common 5′ primer (5′-TGACTAGCGGTACCGGGGTTGCTGCTGTGGAACC) and different 3′ primers (5′-GCACCTGGTTGAGGCCGCGCGCTCGAGCTCTAGTCA for −148/+84luc and 5′-CAAGAGGTCGCGGGGCGCGTCCTCGAGCTCTAGTCA for −148/+57luc). PCR products were isolated and digested with KpnI and XhoI and ligated into KpnI/XhoI digested pGL2Basic (Promega). The plasmid cadG4luc was made by inserting an oligonucleotide containing the Gal4-binding site and HindIII ends (5′-AGCTTAGCCGGAAGACTCTCCTCCGACTA) into the HindIII site downstream of the cad promoter in cad −81/+26luc (21). The plasmid −335 G4cadluc was made by inserting an oligonucleotide containing the Gal4-binding site and XbaI ends (5′-CCGGGAGGCCTCGGAAGACTCTCCTCCGTGC) into the SmaI site upstream of the cad promoter in cad −335/+26luc (22). The expression vectors for the Gal4 fusion proteins, Gal4-Myc, Gal4-USF1, and Gal4-TFE3, and the Gal4 DNA-binding domain alone were gifts of M. Eilers (10), and Gal4-USF2 (G-U2N) was obtained from M. Sawadogo (23). The plasmids dhfrG4 and −375G4dhfr were described previously (24).
Formaldehyde Crosslinking and Immunoprecipitation of Chromatin.
Formaldehyde (Fisher Scientific) was added directly to cell culture media at a final concentration of 1% at 0, 4, 8, or 12 hr after serum addition to serum-starved NIH 3T3 cells or to nonadherent log phase HeLa cells. Fixation proceeded at 22°C for 10 min and was stopped by the addition of glycine to a final concentration of 0.125 M. To harvest NIH 3T3 cells, plates were rinsed with cold PBS, incubated with 5 ml of 0.2× trypsin-EDTA (GIBCO) in PBS, and then scraped. Cells were collected by centrifugation and washed in cold PBS plus 0.5 mM phenylmethylsulfonyl fluoride. HeLa cells were collected by centrifugation and washed as above. Cells were swelled in 5 mM Pipes (pH 8.0), 85 mM KCl, 0.5% NP40, 0.5 mM phenylmethylsulfonyl fluoride, and 100 ng/ml leupeptin and aprotinin, incubated on ice for 20 min, and then dounced. Nuclei were collected by microcentrifugation at 5,000 rpm, resuspended in sonication buffer [1% SDS, 10 mM EDTA, 50 mM Tris⋅HCl (pH 8.1), 0.5 mM phenylmethylsulfonyl fluoride, and 100 ng/ml leupeptin and aprotinin] and incubated on ice for 10 min. Samples were sonicated on ice to an average length of 500–1,000 bp and then microfuged at 14,000 rpm. The chromatin solution was precleared with the addition of Staph A cells for 15 min at 4°C. Before use, Staph A cells were blocked with 1 μg/μl sheared herring sperm DNA and 1 μg/μl BSA for at least 4 hr at 4°C. Precleared chromatin from 2.5 × 107 cells was incubated with 1 μg of affinity-purified rabbit polyclonal antibody (Santa Cruz: anti-Myc sc-764-X, anti-Max sc-765X, anti-E2F4 sc-1082-X), 1 μl of anti-human USF1 antiserum from rabbits (gift of E. Bresnick), or no antibody and rotated at 4°C for 12 hr. Immunoprecipitation, washing, and elution of immune complexes was carried out as described (16). Before the first wash, the supernatant from the reaction lacking primary antibody for each time point was saved as total input of chromatin and was processed with the eluted immunoprecipitates beginning at the crosslink reversal step. Crosslinks were reversed by addition of NaCl to a final concentration of 200 mM, and RNA was removed by addition of 10 μg of RNase A per sample followed by incubation at 65°C for 5 hr. Samples were then precipitated at −20°C overnight by the addition of 2 vol of ethanol and then pelleted by microcentrifugation. Samples were resusupended in 100 μl of TE (10 mM Tris, pH 7.5/1 mM EDTA), 25 μl of 5× proteinase K buffer (1.25% SDS/50 mM Tris, pH 7.5/25 mM EDTA), and 1.5 μl of proteinase K (Boehringer Mannheim) and incubated at 42°C for 2 hr. Samples were extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and then precipitated with 1/10th vol of 3 M sodium acetate (pH 5.3), 5 μg tRNA, and 2 vol of ethanol at −20°C overnight. Pellets were collected by microcentrifugation, resuspended in 30 μl of H2O, and analyzed by using PCR. Total input samples were resuspended in 100 μl of H2O and then diluted 1:100 before PCR. PCR reactions contained 2 μl of immunoprecipitate or diluted total input, 50 ng of each primer, 0.88 mM MgCl2, 2 mM each dATP, dCTP, dGTP, dTTP, 1× Thermophilic buffer (Promega), and 1.25 units Taq DNA polymerase (Promega) in a total volume of 20 μl. After 32–35 cycles of amplification, PCR products were run on a 1.5% agarose gel and analyzed by ethidium bromide staining. (A detailed protocol is available on request.)
RESULTS
Cloning and Characterization of the Murine cad Promoter.
Previously we showed that expression from the hamster cad promoter is growth-regulated and that regulation is dependent on an E box located downstream of the transcription start site. We wished to determine which E box-binding proteins bound to the endogenous cad promoter in different stages of the cell cycle. Mouse NIH 3T3 cells are commonly used when analyzing growth-regulated transcription. However, the mouse cad promoter had not been cloned. Therefore, before analyzing the transcription factor binding in NIH 3T3 cells, it was necessary to clone the murine cad promoter, determine whether expression from the murine cad promoter is growth regulated, and if so, determine whether this regulation requires an E box.
To clone the murine cad promoter, PCR was performed by using primers directed toward two regions which, are conserved between the hamster and human cad promoters (ref. 20 and S. Mac and P.J.F., unpublished results). The sequence of the resulting 659-bp PCR fragment was 85.7% identical to the hamster cad promoter, verifying that the mouse cad promoter had been cloned. As shown in Fig. 1A, the critical promoter elements of the hamster promoter are conserved in the murine cad promoter. Importantly, the E box, which is known to be required for growth-regulated transcription of the hamster promoter, also has been conserved and is present in the same location in the murine gene.
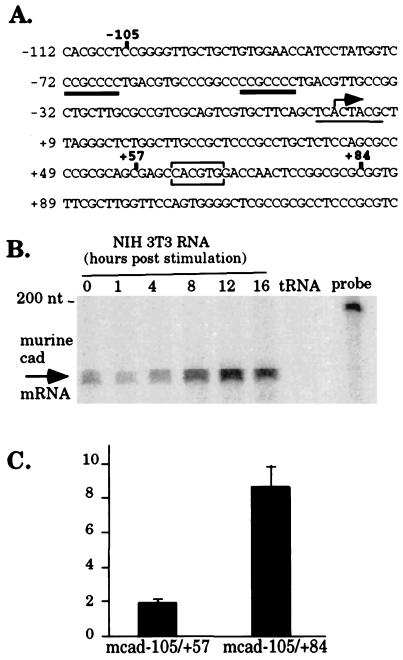
Figure 1.
The mouse cad promoter displays E box-dependent growth-regulated transcriptional activity. (A) Sequence analysis of the mouse cad-promoter region from −112 to +129. Numbering of nucleotides is based on the putative start of transcription at +1 (indicated by the bent arrow). Transcription factor-binding sites, which are conserved between the murine and hamster cad promoters are indicated as follows: Sp1-binding sites are double underlined, the initiator element is underlined, and the E box is boxed. (B) Expression of cad mRNA displays late serum-response kinetics in mouse NIH 3T3 cells. Cytoplasmic RNA (10 μg), harvested at the indicated times after serum stimulation of quiescent NIH 3T3 cells or tRNA (10 μg) was hybridized to a murine cad riboprobe. Unprotected RNA was removed by digestion with RNase A. Protection of murine cad mRNA results in a 154-nt product, as indicated by the arrow. (C) Activated transcription from the mouse cad promoter in S phase requires the E box. Murine cad-promoter sequences from −105 to +84 or from −105 to +57 (indicated in bold in A) were fused upstream of the luciferase cDNA and transiently transfected into NIH 3T3 cells. After transfection, cells were incubated in starvation media (DMEM + 0.5% calf serum) for 48 hr, stimulated with the addition of 10% calf serum for 16 hr (corresponding to S phase of the growth cycle), and harvested for measurement of luciferase activity. The fold induction at 16 hr is reported as the ratio of luciferase activity from serum stimulated cells to the activity of the same promoter construct in serum-starved cells. Average fold induction and standard error was calculated from three independent experiments.
The next step was to determine whether expression from the murine cad promoter is growth regulated and whether this regulation requires the presence of the E box. We examined expression from the endogenous murine cad promoter in synchronized NIH 3T3 cells by using a RNase protection assay. NIH 3T3 cells were serum starved for 48 hr and then harvested for RNA at 0, 1, 4, 8, 12, and 16 hr after serum stimulation. RNA from each time point was incubated with a murine cad riboprobe, and unprotected RNA was removed by digestion with RNase A. As shown in Fig. 1B, the abundance of cad mRNA increases after serum stimulation of NIH 3T3 cells with significant accumulation of cad mRNA appearing at 8 hr after the addition of serum. The peak of cad expression is observed by 12 hr after stimulation, which corresponds to the G1/S phase boundary of the growth cycle (as determined by flow cytometry, data not shown).
To evaluate the contribution of the E box to serum-responsive cad expression, we created murine cad promoter constructs either containing (mcad-105/+84) or lacking the E box (mcad −105/+57). Plasmids containing the cad-promoter fragments cloned upstream of the luciferase cDNA were transiently transfected into NIH 3T3 cells. After transfection, the cells were serum starved for 48 hr, serum stimulated for 16 hr, allowing the cells to enter early S phase, and then assayed for luciferase activity. As shown in Fig. 1C, transcription from the −105/+84 murine cad promoter is increased after serum stimulation and high level promoter activity in S phase is dependent on the presence of the E box.
Myc Binds to the cad Promoter in Vivo After Serum Stimulation of Quiescent Cells.
Having demonstrated that the murine cad promoter displays E box-dependent growth regulation, the next step was to examine protein binding to the endogenous cad promoter at different stages of the growth cycle. Based on previous UV crosslinking results with cells containing 100 copies of the cad gene (16), it was clear that detection of protein binding to diploid copy genes would require a PCR-based assay. However, PCR analysis of UV-crosslinked chromatin was inefficient and not highly reproducible (unpublished data). Therefore, we adapted a reversible formaldehyde-crosslinking procedure previously used to study histone binding in Drosophila to investigate transcription factor binding in mammalian cells (25). Our modified protocol is outlined in Fig. 2.
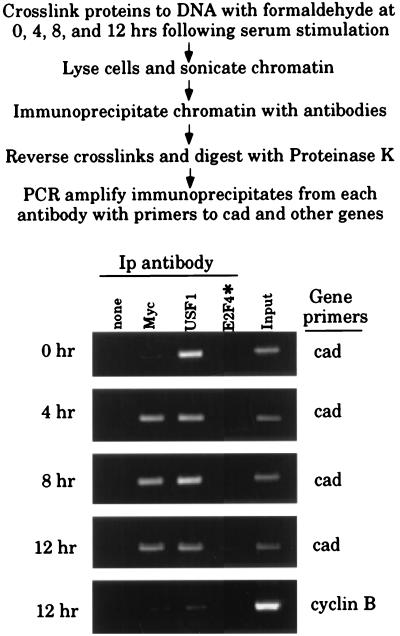
Figure 2.
Myc binds to the cad promoter in vivo after serum stimulation of quiescent cells. Shown is the protocol for the formaldehyde crosslinking and chromatin immunoprecipitation assay used to detect protein binding to single copy genes. Formaldehyde-crosslinked chromatin was prepared from the same number of NIH 3T3 cells that were serum starved (0 hr) or serum starved and then stimulated for 4, 8, or 12 hr. Crosslinked chromatin from each time point was incubated with antibodies to Myc, USF1, or in the absence of antibody (none). In this experiment, E2F4 antibody (highlighted by the asterisk) was only incubated with the 12-hr chromatin. Immunoprecipitates from each antibody were aliquotted and then analyzed by PCR with primers specific for the cad or cyclin B promoters. To verify that at each time point an equivalent amount of chromatin was used in the immunoprecipitations, a sample representing 0.02% of the total input chromatin (input) was included in the PCR reactions.
Our previous UV-crosslinking studies demonstrated that both Myc and USF1 can bind the cad promoter in asynchronously growing cells. However, it is possible that Myc and USF1 are bound to the cad promoter during different phases of the cell cycle. We wanted to determine which factor is bound to the cad promoter in vivo during the peak of cad transcription and whether unique binding patterns are observed for Myc and USF1. To address this question, serum-starved NIH 3T3 cells were treated with formaldehyde at 0, 4, 8, and 12 hr after serum stimulation. The crosslinked chromatin from equivalent numbers of quiescent and stimulated cells was then immunoprecipitated by using antibodies against Myc and USF1. As controls, we included a reaction lacking primary antibody and a reaction that contained E2F4 antibody. E2F4 is a nuclear transcription factor abundant in proliferating cells, which does not have a binding site in the cad promoter; therefore, this antibody should not immunoprecipitate cad from crosslinked chromatin. Each of the antibodies was shown by Western blot analysis to detect their cognate protein in NIH 3T3 nuclear extract (data not shown). After immunoprecipitation and reversal of the crosslinks, enrichment of the endogenous cad-promoter fragment in each sample was monitored by PCR amplification using primers specific for the cad promoter. As shown in Fig. 2, the pattern of Myc and USF1 binding differ on the cad promoter. Anti-Myc immunoprecipitates from quiescent chromatin contain nearly undetectable levels of cad-promoter fragment; however, at 4, 8, and 12 hr after serum stimulation, the same antibody readily immunoprecipitates cad. In contrast, the USF1 antibody immunoprecipitated the cad promoter from both quiescent and serum-stimulated cells. Identical results were obtained when PCR reactions were amplified for 14 fewer cycles and analyzed by Southern blotting, verifying that the PCR amplification was not significantly beyond the linear range (data not shown). Enrichment for cad promoter containing fragments is dependent on Myc and USF1 protein binding to cad because an antibody against E2F4 does not immunoprecipitate cad-promoter fragments. Additionally, binding detected at the cad promoter is specific, since antibodies against Myc and USF1 do not enrich for cyclin B-promoter fragments to the same level as for cad-promoter fragments (Fig. 2 Lower). These results demonstrate that after serum stimulation of quiescent cells the cad promoter is bound by Myc and that USF1 binds to cad in both quiescent and proliferating cells.
Transcriptional Activation Is Determined by Activation Domain Specificity, Not by DNA-Binding Specificity.
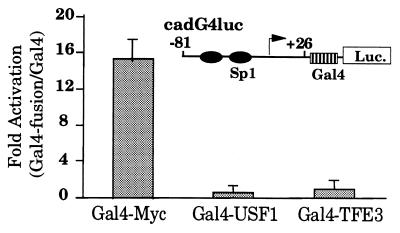
The results obtained by using formaldehyde crosslinking indicate that both Myc and USF1 bind to the cad promoter. However, we show that USF1 is bound to cad in quiescent cells, when promoter activity is very low, suggesting that target gene specificity is not achieved at the level of DNA binding but perhaps at the level of transactivation. To test this hypothesis, we used a cad promoter–reporter construct, cadG4luc, containing a Gal4-binding site inserted downstream of the cad-81/+26 minimal promoter. We have shown previously that an E box cloned at this position will confer growth regulation to the minimal cad promoter (15). We then monitored the ability of the transactivation domains of three different E box-binding proteins to increase transcriptional activity from the cad promoter. NIH 3T3 cells were cotransfected with the cadG4luc reporter construct and an expression vector for the Gal4 DNA-binding domain or the Gal4 DNA-binding domain fused to the transactivation domains of Myc, USF1, or TFE3. As shown in Fig. 3, the cad promoter can be activated by the transactivation domain of Myc but not USF1 or TFE3. We also tested the ability of Gal4-USF2, another member of the USF protein family, to activate cad expression and obtained similar results as seen for Gal4-USF1 (data not shown). However, we did find that all of the fusion proteins were able to activate expression from a synthetic reporter plasmid, containing multiple Gal4 sites cloned upstream of a TATA box, to levels observed in other studies (data not shown). Together these results demonstrate that Myc, USF1, USF2, and TFE3 are not equivalent in their ability to activate cad transcription. Therefore, differential activation of the cad promoter by E box-binding proteins is determined at the level of protein–protein interactions and not at the level of DNA binding.
Figure 3.
Transcription from the cad promoter is activated by the transactivation domain of Myc but not USF1. Asynchronously growing NIH 3T3 cells were transiently cotransfected with 0.5 μg of cadG4luc reporter plasmid and 2.5 μg of Gal4-Myc, Gal4-USF1, and Gal4-TFE3 expression plasmid. The cells were harvested for luciferase activity at 48 hr after transfection. For each experiment, activation of the cad reporter by the Gal4-fusion proteins was normalized to the activity of the Gal4 DNA-binding domain on the same reporter. Average activation and SEM was calculated from seven to 10 independent experiments by using multiple DNA preparations.
Transcriptional Activation by Myc Is Regulated by the Position of Myc Binding.
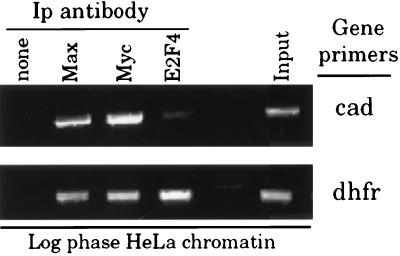
During the course of these formaldehyde crosslinking studies, we established that Myc binding occurs at the cad promoter, which contains a consensus E box element. However, there exists extensive evidence through both in vitro- and in vivo-binding studies that Myc can bind to many types of nonconsensus E box elements such as CATGTG, CACCTG, CATGCG, CACGCG, CAACGTG, and CACGAG (14, 26). Statistically, consensus and these nonconsensus E boxes should be distributed throughout the genome at a frequency of ≈1/800 bp (14). To evaluate Myc activity at nonconsensus E box elements, we examined the human dhfr promoter. This promoter contains two nonconsensus elements (CACCTG) positioned between −378 and −364, which have been proposed to bind Myc in vitro (27). The dhfr promoter also has overlapping E2F-binding sites centered at the transcriptional start site. By using formaldehyde-crosslinked HeLa cell chromatin, we examined Myc, Max, and E2F4 binding to the human dhfr promoter. In the same experiment, we examined binding to the human cad promoter, which has two consensus E box elements (S. Mac and P.J.F, unpublished results). As expected, E2F4 binding is detected only on the dhfr promoter (Fig. 4). However, Myc and Max binding is detected on both the cad and dhfr promoters, indicating that Myc can bind to nonconsensus E boxes in vivo as well as in vitro.
Figure 4.
Myc binds to the human cad and dhfr promoters in vivo. Crosslinking analysis of Myc, Max, and E2F4 binding at the human cad and dhfr promoters in log phase HeLa cells. Equivalent amounts of formaldehyde-crosslinked chromatin from asynchronously growing HeLa cells was incubated with antibodies to Myc, Max, E2F4, or in the absence of antibody (none). Immunoprecipitation of the human cad and dhfr promoters by each antibody was analyzed in parallel PCR reactions by using primers specific to the cad or dhfr promoters. The input sample (input) contains 0.02% of the total input chromatin as PCR template for each set of primers.
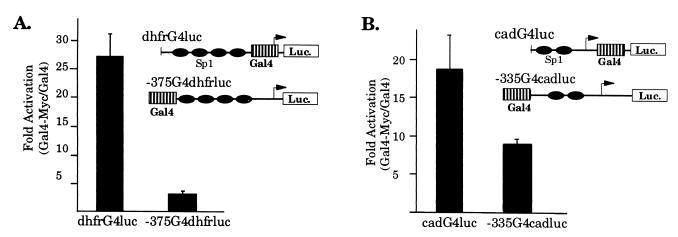
Next, we examined the effect of binding position on Myc transactivation activity on the dhfr and cad promoters. To evaluate Myc activity on dhfr expression, we used a mouse dhfr promoter–reporter construct containing a Gal4-binding site cloned at −375, similar to the location of the nonconsensus E boxes in the human dhfr promoter. As shown in Fig. 5A, cotransfection of this reporter with Gal4-Myc does not activate expression from the dhfr promoter. However, when the Gal4 site is moved to +1 in the dhfr promoter, Gal4-Myc robustly activates dhfr expression. To examine whether Myc activity is also position dependent in the context of the cad promoter, similar Gal4-Myc studies were performed by using a cad promoter–reporter plasmid containing a Gal4 site cloned at −335. As shown in Fig. 5B, changing the Gal4-Myc-binding site to a distal upstream position in the cad promoter results in only a twofold difference in Myc activity suggesting that transactivation of cad, unlike dhfr, is relatively insensitive to the position of Myc binding.
Figure 5.
Transcriptional activation by Myc is dependent on the context of Myc binding. (A) The transactivation domain of Myc cannot activate expression from the dhfr promoter from the distal upstream position. Asynchronous NIH 3T3 cells were transiently cotransfected with 2.5 μg of dhfrG4luc or −375G4dhfrluc reporter and 5.0 μg of Gal4-Myc expression plasmid. Cells were harvested for luciferase activity 48 hr after transfection. For each experiment, activation of the dhfr reporter by Gal4-Myc was normalized to the activity of the Gal4 DNA-binding domain on the same reporter. Data presented was obtained from five to seven independent experiments, using multiple DNA preparations. (B) Activation of cad expression is less sensitive to the position of Myc binding. Asynchronous NIH 3T3 cells were transiently cotransfected with 0.5 μg of cadG4luc or −335cadG4luc reporter and 2.5 μg of Gal4-Myc expression plasmid. Cells were harvested and analyzed as described above. Data presented was obtained from eight to nine independent experiments.
DISCUSSION
To better understand how target gene specificity is achieved between members of the bHLHzip family, we have analyzed the in vivo-binding specificity of Myc and USF1 and investigated the functional consequences of this binding on gene expression. Adaptation of a formaldehyde-crosslinking and immunoprecipitation protocol has allowed us to detect binding of Myc, USF1, and other nuclear transcription factors to the promoters of cellular genes within the context of an intact cell. Our in vivo-crosslinking results indicate that the cad promoter is bound by USF1 at all stages of the growth cycle and that Myc binding parallels the growth-regulated increase in Myc protein after stimulation of quiescent cells. Because USF1 is bound constitutively to the cad promoter even in G0 when cad expression is very low, we propose that this binding is nonproductive. Our model is supported by the observation that the transactivation domain of Myc, but not USF1, activates transcription when bound to the cad promoter in the normal downstream position. It remains unclear whether Myc binding displaces USF1 as cells progress through the growth cycle. Because our crosslinking assay is based on large populations of cells, further analysis of cad gene expression in single cells is required to address this question.
We have detected Myc binding to regions of DNA, which contain both consensus and nonconsensus E box motifs. In addition to dhfr, we observed Myc bound to very large chromatin fragments containing the B-myb and thymidine kinase genes (unpublished observations). These results are in agreement with the findings of Grandori et al. (14) who demonstrated that in vivo Myc can bind many nonconsensus E box sequences, which are distributed frequently throughout the genome. One caveat of the previous study is that binding site selection was evaluated in cells that overexpressed Myc. Our results provide evidence that under physiological conditions, Myc binding also is widely distributed. The finding that large chromatin fragments containing nonconsensus E boxes can be immunoprecipitated by using Myc antibodies underscores the importance of using chromatin sonicated to a small size (<1 kb) to evaluate binding at a specific promoter region. The broad binding specificity we observed for Myc is not common to all transcription factors tested. For example, we have shown that E2F-binding sites occur less frequently in the genome and that different E2F target genes display different patterns of binding of E2F family members (data not shown). Our studies of mammalian transcription factors are in agreement with those using Drosophila as a model system. Two distinct classes of Drosophila DNA-binding proteins have been characterized in vivo; those with a broad binding pattern such as the homeodomain proteins eve and ftz, and those which display a narrow range of binding targets such as zeste (28).
In light of the frequent occurrence of Myc-binding sites, it is not likely that Myc activates expression of all genes to which it is bound. We propose that Myc target gene selectivity is achieved through a post-DNA-binding mechanism. In support of this model, our Gal4 fusion protein experiments indicate that Gal4-Myc cannot transactivate all cellular promoters tested. We show that Gal4-Myc does not activate expression of the dhfr promoter when it is bound far upstream of the transcriptional start site where both canonical and noncanonical E box elements are normally found in the hamster and human promoters. It has been suggested that Myc can activate transcription in a position independent manner, using an artificial promoter construct (29). However, it is likely that endogenous cellular promoters may show different extents of position dependence. Accordingly, we show that transactivation of the cad promoter is relatively insensitive to the position of Myc binding. Our results strongly suggest that Myc binding may not always correlate with Myc transcriptional activity. Although much of the bound Myc in the cell may have little influence on promoter activity, it is possible that Myc binding to certain sites may result in transcriptional repression. Thus, assays that are based on Myc function should prove more useful in the search for Myc target genes than those which rely solely on Myc binding.
In summary, our studies suggest that cad is a Myc target gene. cad is bound by Myc in vivo and the Myc transactivation domain is functional when bound near the cad core promoter. In addition, studies using cells that are nullizygous for c-myc show a significant reduction in cad mRNA expression after serum stimulation (A. Bush and M. Cole, unpublished results). Although the timing of Myc binding to the cad promoter correlates with the pattern of Myc protein expression after serum stimulation (15), there is a lag between the binding of Myc in early G1 and cad activation in late G1. This result suggests that a post-DNA-binding modification is necessary to increase Myc transcriptional activity. The transactivation domain of Myc has been shown to be a target of phosphorylation by many protein kinases such as mitogen-activated protein kinase (30), mitogen-activated protein kinase 5 (31), and cyclin A-cyclin-dependent kinase (32). Further evaluation of the contribution of these kinases to Myc-mediated activation of cad expression is in progress. Finally, we propose that our approach, which uses a formaldehyde-crosslinking technique to examine in vivo protein binding in combination with reporter gene assays, will be applicable to other investigators in their attempts to determine which member of a family of transcription factors regulates a particular cellular target gene.
Acknowledgments
We are grateful to David Allis and Richard Treisman for sharing their formaldehyde-crosslinking protocols. We thank Martin Eilers and Michele Sawadogo for providing the Gal4 fusion plasmids and Emery Bresnick for anti-USF1 rabbit antiserum. This work was supported in part by Public Health Service Grants CA07175, CA45240, CA09135, and CA09681 from the National Institutes of Health (to P.F.) and a Hilldale Undergraduate Research Fellowship (to J.G.).
ABBREVIATIONS
- dhfr
dihydrofolate reductase
- bHLHzip
basic helix–loop–helix leucine zipper
- mcad
mouse cad
- USF
upstream stimulatory factor
Footnotes
References
- 1.Blackwood E M, Eisenman R N. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 2.Ayer D E, Kretzner L, Eisenman R N. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 3.Zervos A S, Gyuris J, Brent R. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 4.Hurlin P J, Queva C, Eisenman R N. Genes Dev. 1997;11:44–58. doi: 10.1101/gad.11.1.44. [DOI] [PubMed] [Google Scholar]
- 5.Gregor P D, Sawadogo M, Roeder R G. Genes Dev. 1990;4:1730–1740. doi: 10.1101/gad.4.10.1730. [DOI] [PubMed] [Google Scholar]
- 6.Beckmann H, Su L, Kadesch T. Genes Dev. 1990;4:167–179. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- 7.Carr C S, Sharp P A. Mol Cell Biol. 1990;10:4384–4388. doi: 10.1128/mcb.10.8.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bello-Fernandez C, Packham G, Cleveland J L. Proc Natl Acad Sci USA. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tobias K E, Shor J, Kahana C. Oncogene. 1995;11:1721–1727. [PubMed] [Google Scholar]
- 10.Desbarats L, Gaubatz S, Eilers M. Genes Dev. 1996;10:447–460. doi: 10.1101/gad.10.4.447. [DOI] [PubMed] [Google Scholar]
- 11.Benvenisty N, Leder A, Kuo A, Leder P. Genes Dev. 1992;6:2513–2523. doi: 10.1101/gad.6.12b.2513. [DOI] [PubMed] [Google Scholar]
- 12.Jones R M, Branda J, Johnston K A, Polymenis M, Gadd M, Rustgi A, Callanan L, Schmidt E V. Mol Cell Biol. 1996;16:4754–4764. doi: 10.1128/mcb.16.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galaktionov K, Chen X, Beach D. Nature (London) 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 14.Grandori C, Mac J, Siebelt F, Ayer D E, Eisenman R N. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- 15.Miltenberger R J, Sukow K, Farnham P J. Mol Cell Biol. 1995;15:2527–2535. doi: 10.1128/mcb.15.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyd K E, Farnham P J. Mol Cell Biol. 1997;17:2529–2537. doi: 10.1128/mcb.17.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo X, Sawadogo M. Proc Natl Acad Sci USA. 1996;93:1308–1313. doi: 10.1073/pnas.93.3.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bendall A J, Molloy P L. Nucleic Acids Res. 1994;22:2801–2810. doi: 10.1093/nar/22.14.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slansky J E, Li Y, Kaelin W G, Farnham P J. Mol Cell Biol. 1993;13:1610–1618. doi: 10.1128/mcb.13.3.1610. (correction, 13, 7201). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farnham P J, Kollmar R. Cell Growth Differ. 1990;1:179–189. [PubMed] [Google Scholar]
- 21.Kollmar R, Sukow K A, Sponagle S K, Farnham P J. J Biol Chem. 1994;269:2252–2257. [PubMed] [Google Scholar]
- 22.Kollmar R. Ph.D. thesis. Madison, WI: University of Wisconsin; 1993. [Google Scholar]
- 23.Luo X, Sawadogo M. Mol Cell Biol. 1996;16:1367–1375. doi: 10.1128/mcb.16.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fry C J, Slansky J E, Farnham P J. Mol Cell Biol. 1997;17:1966–1976. doi: 10.1128/mcb.17.4.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solomon M J, Larsen P L, Varshavsky A. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 26.Blackwell T K, Huang J, Ma A, Kretzner L, Alt F, Eisenman R, Weintraub H. Mol Cell Biol. 1993;13:5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mai S, Jalava A. Nucleic Acids Res. 1994;22:2264–2273. doi: 10.1093/nar/22.12.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walter J, Dever C A, Biggin M D. Genes Dev. 1994;8:1678–1692. doi: 10.1101/gad.8.14.1678. [DOI] [PubMed] [Google Scholar]
- 29.Packham G, Bello-Fernandez C, Cleveland J L. Cell Mol Biol Res. 1995;40:699–706. [PubMed] [Google Scholar]
- 30.Alvarez E, Northwood I C, Gonzalez F A, Latour D A, Seth A, Abate C, Curran T, Davis R J. J Biol Chem. 1991;266:15277–15285. [PubMed] [Google Scholar]
- 31.English J M, Pearson G, Baer R, Cobb M H. J Biol Chem. 1998;273:3854–3860. doi: 10.1074/jbc.273.7.3854. [DOI] [PubMed] [Google Scholar]
- 32.Hoang A T, Lutterbach B, Lewis B C, Yano T, Chou T Y, Barrett J F, Raffeld M, Hann S R, Dang C V. Mol Cell Biol. 1995;15:4031–4042. doi: 10.1128/mcb.15.8.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]