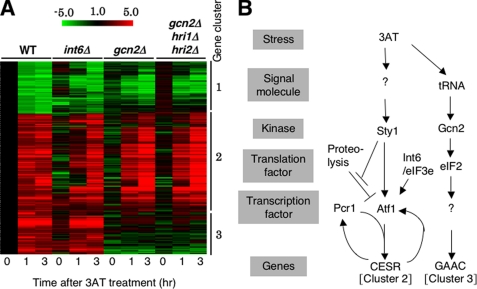
FIGURE 2.
Microarray studies. A, strains used in Fig. 1B were cultured in EMM-C and treated with 30 mm 3AT for the indicated times, followed by RNA extraction and cDNA microarray analysis (27). Shown is the clustering of genes whose transcripts were changed ≥2-fold by 3AT-induced starvation in wild-type cells. Red/green scale bar at the top indicates the fold changes in transcription. Numbers beside the clustering pattern indicate the groups of coregulated genes. B, model for 3AT-induced histidine starvation response pathways in fission yeast. 3AT-induced histidine starvation creates at least two signals that activate the indicated signal transduction pathways. In the first pathway, histidine starvation activates Sty1 MAPK via unidentified signaling molecule(s) (denoted by “?”) by an unknown mechanism (see “Discussion”). Active Sty1 inhibits proteolysis of Atf1 (stopped bars) and also activates transcriptional function of Atf1 (a vertical arrow) (21). Atf1 and Pcr1 together activate CESR (cluster 2 in panel A). Atf1-dependent transcription of atf1+ and pcr1+ (Fig. 5, A and B) may help to enhance CESR transcription (curved arrows). Here we suggest that Int6/eIF3e promotes Atf1 protein translation (and stability) (an arrow). In the second pathway, histidine starvation activates Gcn2 and downstream GAAC response (cluster 3 in panel A) by the mechanism characterized for S. cerevisiae GAAC (13). “?” under Gcn2 indicates the unidentified transcription factor, which would be analogous to Gcn4p in S. cerevisiae.