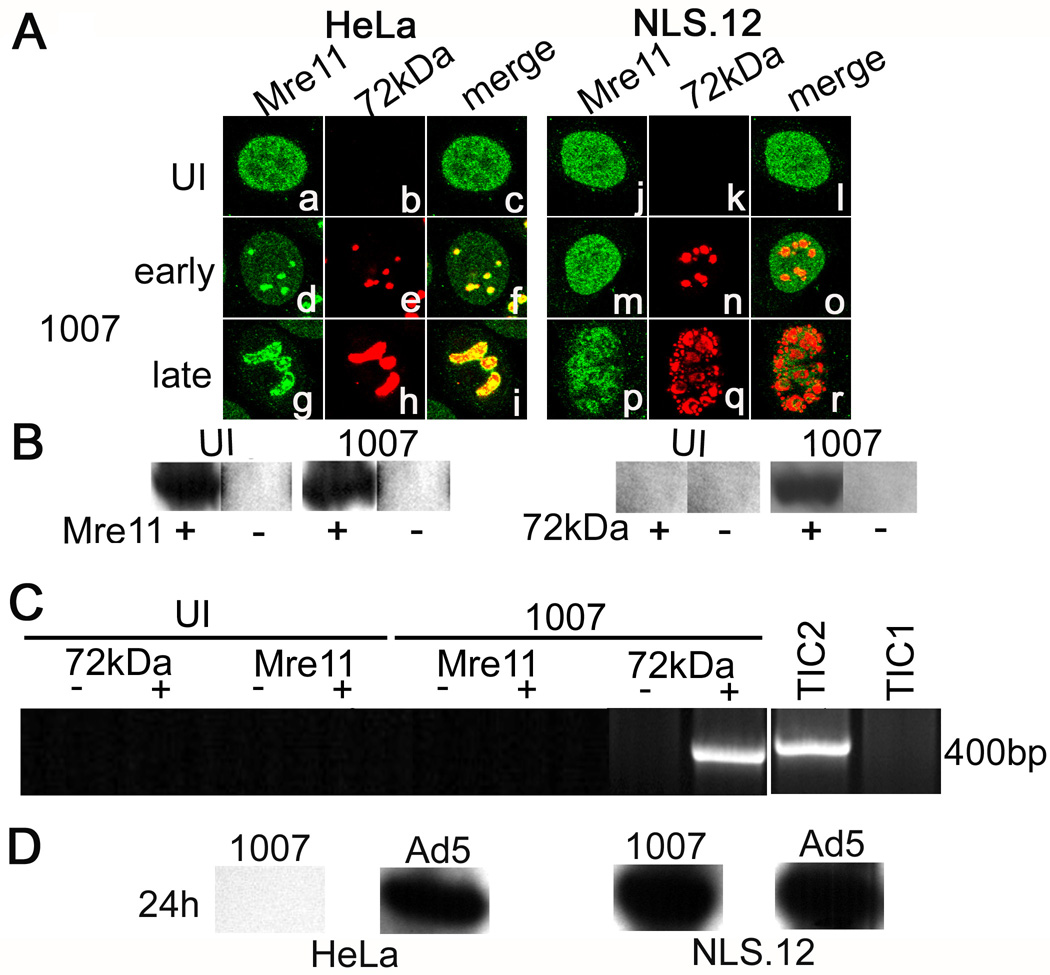
Figure 4. Nbs1 is important for Mre11 localization in E4 mutant replication centers and Mre11 binding to viral DNA.
(A) HeLa and NLS.12 cells were infected with H5dl1007 for 10 h (early) and 24 h (late) at 3 FFU/cell. Confocal microscopy was used to analyze the distribution of host Mre11 (panels a,d,g,j,m,p) and viral E2-72kDa (panels b,e,h,k,n,q) proteins in uninfected and infected cells. Merged images of these staining patterns are shown in panels c, f, i, and l, o, r. ChIP experiments were performed as described in Fig. 1 using samples prepared from NLS.12 cells that were uninfected or infected with H5dl1007 for 12h at 30FFU/cell. (B) Western blotting was performed to confirm immunoprecipitation of Mre11 and E2-72kDa from chromatin prepared from H5dl1007 infected and uninfected NLS.12 cells, in preparation for chIP analysis. Lanes from samples that were immunoprecipitated with specific Ab or mock immunoprecipitated are labeled (+) and (−), respectively. (C) PCR amplification using primers specific to the E1b region was performed on UI and H5dl1007 infected samples prepared at 12 hpi and immunoprecipitated with Mre11 or E2-72kDa. PCR products were analyzed as described in Fig. 1. Lanes from samples that were immunoprecipitated with specific Ab or mock immunoprecipitated in parallel are labeled (+) and (−), respectively. Total input chromatin samples from uninfected (TIC1) and H5dl1007-infected (TIC2) cells were included to indicate input DNA levels. The chIP experiments were performed twice with similar results. (D) Levels of viral DNA synthesis in HeLa and NLS.12 cells infected with Ad5 and H5dl1007 at 3FFU/cellwere quantified by Southern analysis of 10µg of Eco RI digested total DNA prepared at 24 hpi. The C fragment from the DNA digestion was used for comparison between Ad5 and H5dl1007.