Figure 4. Acm1 localization during the cell cycle is controlled by Cdk1.
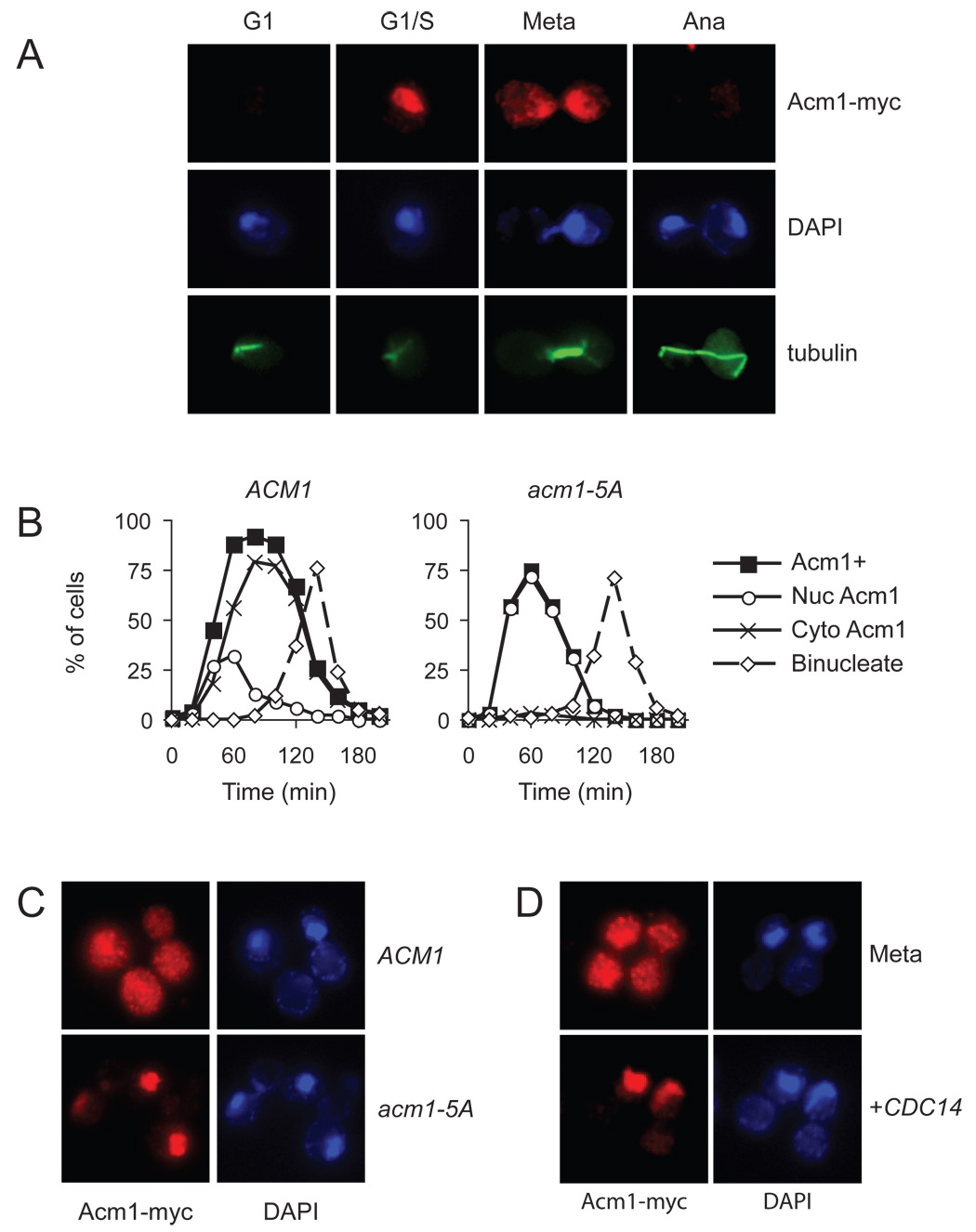
(A) Asynchronous ACM1-myc9 cells were processed for immunofluorescence analysis with anti-Myc and anti-tubulin antibodies, and stained with DAPI for detection of DNA. Cell-cycle position was determined on the basis of cell and spindle morphology and the separation of DNA masses.
(B) ACM1-myc9 and acm1-5A-myc9 cells were arrested in G1 with alpha factor and released into a synchronous cell cycle. Samples were collected every 20 min for analysis of budding, binucleate formation and the presence of Acm1-myc immunofluorescence. Acm1-positive cells were further characterized to determine if Acm1 was exclusively nuclear (Nuc Acm1) or dispersed throughout the cell (Cyto Acm1). For clarity, the time course of budding is not shown here but was very similar to that shown in Figure 5C.
(C) ACM1-myc9 and acm1-5A-myc9 cells were arrested in mitosis by nocodazole treatment, and immunofluorescence performed to assess Acm1 localization.
(D) ACM1-myc9 cells, carrying an integrated plasmid with CDC14 under the control of the GAL1-10 promoter, were blocked in mitosis by nocodazole treatment under noninducing conditions, followed by immunofluorescence analysis of Acm1 localization (top). CDC14 expression was induced by addition of galactose to the culture medium and immunofluorescence performed to assess Acm1 localization (bottom).
