Summary
When performing association studies in populations that have not been the focus of large-scale investigations of haplotype variation, it is often helpful to rely on genomic databases in other populations for study design and analysis – such as in the selection of tag SNPs and in the imputation of missing genotypes. One way of improving the use of these databases is to rely on a mixture of database samples that is similar to the population of interest, rather than using the single most similar database sample. We demonstrate the effectiveness of the mixture approach in the application of African, European, and East Asian HapMap samples for tag SNP selection in populations from India, a genetically intermediate region underrepresented in genomic studies of haplotype variation.
Introduction
High-resolution haplotype maps in populations of European, West African, and East Asian descent provide a basis for efficiently selecting single-nucleotide polymorphisms (SNPs) for use in genetic association studies (Hinds et al., 2005, The International HapMap Consortium, 2005, 2007). Each of these “tag SNPs” is generally chosen to have a high degree of linkage disequilibrium (LD) with many of its neighbors, so that relatively small numbers of tag SNPs genotyped in an association study can capture patterns of genetic variation over broad regions of the genome.
Because dense haplotype maps currently exist only in a relatively small number of populations, tag SNPs for most populations are usually chosen based on data from three groups in the International Haplotype Map Project – European Americans (CEU), Chinese from Beijing and Japanese from Tokyo (CHB+JPT), and Yoruba from Ibadan (YRI). Typically, tag SNPs for a given “target” population are selected based on data from the most similar of the “donor” populations in the HapMap project. In most target populations, patterns of genetic variation can be adequately captured with tag SNPs chosen by this approach (Mueller et al., 2005, Conrad et al., 2006, de Bakker et al., 2006a, 2006b, González-Neira et al., 2006, Huang et al., 2006, Lim et al., 2006, Mahasirimongkol et al., 2006, Montpetit et al., 2006, Ribas et al., 2006, Smith et al., 2006, Stankovich et al., 2006, Willer et al., 2006, Gu et al., 2007, Marvelle et al., 2007, Service et al., 2007). Thus, tag SNPs chosen based on data from the HapMap samples are “portable” to most other populations.
Despite the general success of data from the HapMap Project in tag SNP selection, two groups of populations have been identified in which improvements in tagging procedures may have some potential to increase the effectiveness of tag SNP association studies. One of these groups is sub-Saharan African populations, who have considerably lower levels of LD than other populations (Reich et al., 2001, Gabriel et al., 2002, Tishkoff & Kidd, 2004, Hinds et al., 2005, The International HapMap Consortium, 2005, Sawyer et al., 2005, Conrad et al., 2006) and who therefore require more tag SNPs to attain the same genomic coverage as can be obtained elsewhere. The other group consists of intermediate-LD non-African populations who are genetically distant from populations in the HapMap (Conrad et al., 2006, Johansson et al., 2007, Roy et al., 2008). Such populations – found mainly in parts of Eurasia far from HapMap locations – do not benefit either from the relative ease of identifying tag SNPs in high-LD populations using almost any low- or intermediate-LD donor sample, or from the boost in tag performance supplied by a close genetic relationship to a HapMap population.
To improve the effectiveness of tag SNPs in intermediate-LD non-African populations, we have devised a strategy for tag SNP selection based on mixtures of the HapMap CEU, CHB+JPT, and YRI samples. We construct mixture datasets containing phased haplotypes from the three samples, with specified fractions in the mixture being drawn from CEU, CHB+JPT, and YRI. Tag SNPs are identified from the mixed sample, and the mixture fractions are varied to find values that in a specified non-HapMap population maximize the proportion of non-tag SNPs that exceed a linkage disequilibrium cutoff with at least one tag SNP (“proportion of variation tagged”, or PVT (Conrad et al., 2006)).
For investigating the mixture approach, we have used a dataset of 2,810 SNPs spread across 36 genomic regions previously genotyped in a diverse worldwide collection of 53 populations (Conrad et al., 2006), augmented by similar data on two populations from India, Bengalis and Tamilians. These linguistically defined groups were chosen from a larger survey of Indian genetic variation (Rosenberg et al., 2006) to represent parts of India distant from other places in which haplotype variation has previously been more extensively studied. India has been largely omitted from genomic LD studies, and due to its intermediate location between Europe and East Asia, SNP variation in Indian groups is expected to be imperfectly captured by any single HapMap sample (Roy et al., 2008). Thus, use of mixtures may have some potential for improving the prospects for genetic association studies in Indian populations.
Materials and Methods
Samples
We studied genotypes of 957 unrelated individuals from 55 populations worldwide – 927 individuals from the HGDP-CEPH cell line panel (Cann et al., 2002) and 30 individuals who had previously been included in an investigation of microsatellite variation in Indian populations (Rosenberg et al., 2006). These 30 individuals included 15 Tamilians from the state of Tamil Nadu in southern India and 15 Bengalis with ancestry in the region that before 1948 was the eastern Indian state of Bengal. Two Bantu populations grouped together by Conrad et al. (2006) were analyzed separately here.
Genotype data
Individuals were genotyped using the Illumina BeadLab 1000 platform (GoldenGate genotyping), for 3,024 SNPs spanning 36 genomic regions: 16 from chromosome 21, 16 scattered across the remaining autosomes, and 4 from the non-pseudoautosomal X chromosome. Each region was designed to be centered around a high-density “core” of 60 SNPs, with 12 flanking SNPs at lower density extending from the core at each end. The 30 Indian individuals were genotyped together with 160 African individuals (unpublished data) and 2 control samples. Raw traces from the genotyping assays of 1,248 samples were combined for genotype calling; thus, scoring of genotypes was performed for the 192 samples together with rescoring of genotypes for 1,056 samples studied by Conrad et al. (2006) – the 927 individuals on which Conrad et al. (2006) focused, 121 HGDP-CEPH individuals and 4 HGDP-CEPH duplicate samples genotyped but discarded by Conrad et al. (2006), and 4 controls. Rescored genotypes were used in place of data taken directly from Conrad et al. (2006), resulting in a small number of genotype changes. Of the 2,834 high-quality SNPs studied by Conrad et al. (2006) in 927 individuals, a subset of 2,810 SNPs was investigated in the current study (see below). Considering these 2,810 SNPs, 159 diploid genotypes changed upon rescoring. Thus, excluding individual/genotype combinations with missing data (either in Conrad et al. (2006) or in the rescored data), the discrepancy rate was ~6×10−5.
Quality checks were performed in a collection of 1,107 individuals – the 30 Indians, 150 of the 160 Africans, and the 927 individuals from Conrad et al. (2006). In this set of individuals, the missing data rate was calculated for each SNP, and each SNP was tested for monomorphism. Finally, as severe Hardy-Weinberg disequilibrium can reflect genotyping error, each SNP was tested for Hardy-Weinberg disequilibrium in two relatively unstructured populations – the collection of 30 Indians and a collection of 36 Borana and Iraqw individuals included among the Africans. These groupings were chosen because they had sufficient sample size for Hardy-Weinberg tests but did not have sufficient population structure to introduce substantial Hardy-Weinberg disequilibrium. To be excluded on the grounds of Hardy-Weinberg disequilibrium, a SNP needed to have (1) at least three copies of both alleles in both groups (Indians and Borana/Iraqw), (2) a Yates-corrected chi-squared test statistic greater than 4 in both groups, and (3) a Yates-corrected chi-squared test statistic greater than 8 in at least one of the groups.
The total number of SNPs discarded was 214, in the following categories: (1) 94 SNPs that failed genotyping both in this study and in Conrad et al. (2006); (2) 21 SNPs that failed genotyping in this study but not in Conrad et al. (2006); (3) 21 SNPs that failed genotyping in Conrad et al. (2006) but not in this study; (4) 11 SNPs that failed genotyping in this study, that did not fail genotyping in Conrad et al. (2006) but that were among 75 SNPs discarded from Conrad et al. (2006) for other reasons including missing data, monomorphism, or Hardy-Weinberg disequilibrium; (5) 64 SNPs that did not fail genotyping in this study, but that were discarded from Conrad et al. (2006) due to missing data, monomorphism, or Hardy-Weinberg disequilibrium; (6) 3 SNPs that were used in Conrad et al. (2006), but that failed quality checks in the newly genotyped individuals. One of the three SNPs had excess missing data (>10%). A second of these SNPs was polymorphic in Conrad et al. (2006), but due to changes in genotype calls, it became monomorphic. The third SNP was excluded on the basis of Hardy-Weinberg disequilibrium.
The exclusions produced a high-quality dataset of 2,810 polymorphic SNPs, each of which was among the 2,834 SNPs studied by Conrad et al. (2006). Our final dataset utilized these 2,810 SNPs in 957 individuals – the 30 Indians and the 927 individuals from Conrad et al. (2006). The missing data rate in the cleaned data for the 957 individuals was 0.07% (0.02% in the 30 Indian individuals).
Haplotype phasing
Haplotype phasing utilized fastPHASE 1.0 (Scheet & Stephens, 2006) following the same approach as that of Conrad et al. (2006). Phasing was performed on a dataset consisting of the 30 Indian individuals, the 927 individuals studied by Conrad et al. (2006), and 150 of the 160 additional African individuals. As in Conrad et al. (2006), phasing used separate parameter sets for major geographic regions, placing the Indians with Central/South Asia and grouping the newly genotyped Africans with HGDP-CEPH Africans. This separation by regions during phasing was found to reduce error rates in previous analysis (Conrad et al., 2006). During the phasing of the 1107 individuals, without employing any reference individuals with known haplotypes, fastPHASE was also used to impute all missing genotypes. Low error rates in phasing and missing data imputation (Conrad et al., 2006, Scheet & Stephens, 2006, Andrés et al., 2007, Landwehr et al. 2007, Li & Li 2007, Roberts et al. 2007, Yu & Schaid, 2007) suggest that use of phased haplotypes from fastPHASE is generally suitable in analyses of the r2 linkage disequilibrium statistic.
Combining data with the HapMap
For various analyses, we used the SNPs that overlapped with SNPs in the HapMap Phase II data (release 19) for 32 regions (X-chromosomal regions 23–26 were excluded). A total of 1,853 SNPs overlapped with the HapMap for the 32 regions. Phased haplotypes from 210 individuals in the HapMap – the 60 parents in CEU parent/parent/offspring trios, the 60 parents in YRI trios, and the 90 individuals in the combined CHB+JPT group – were taken directly from the data of Conrad et al. (2006), restricting attention to SNPs not among the 214 of 3024 that were excluded above. One SNP from the Conrad et al.(2006) study, rs12123995, had opposite alleles aligned in the HGDP-CEPH and HapMap datasets; for the current study, the polarity of this SNP was corrected. Thus, 1,853 SNPs from 1,167 individuals (30 Indian individuals, 927 HGDP-CEPH individuals and 210 HapMap individuals) were retained for analyzing tag SNP performance. As in Conrad et al. (2006), the CHB and JPT HapMap samples were combined into one 90-individual panel for all analyses (CHB+JPT).
Linkage disequilibrium
LD was measured using the correlation coefficient r2 for all pairs of SNPs with minor allele frequency (MAF) at least c (where c is a cutoff value in [0,1]). Separately for each population, we computed the mean r2 and the mean distance between pairs of SNPs for all SNP pairs within bins of size b. For example, a bin centered on distance x contains all pairs of SNPs separated by a distance in the interval (x−b/2, x+b/2]. Several choices of c (0, 0.05 and 0.1) and b (1 kb, 3 kb, 6 kb and 10 kb) were tested, and the choices of c and b had relatively little effect on the observed LD patterns. For these computations, we used all core SNPs excluding X-chromosomal regions 23–26 (1,801 SNPs).
Haplotype sharing with the HapMap
For each population we used the ϕ statistic (Conrad et al., 2006) to compute the fraction of haplotypes common in a population that are also common in the HapMap. This approach determines the sample size-corrected number of distinct haplotypes common in each of a pair of populations, as a fraction of the sample size-corrected number of distinct haplotypes common in the population from the pair designated as the “donor.” We found that the choice of cutoff for the definition of “common” (>0.01, >0.05, >0.1) had little effect on the computation. Because the smallest sample size among the 55 populations is 12 chromosomes (San), we used g=12 in the rarefaction-based evaluations of the number of distinct haplotypes. Of 1,853 autosomal SNPs overlapping with the HapMap, 1,309 are core SNPs. These 1,309 SNPs were used for the computations of ϕ, and the two components of regions 30–32 with gaps were each treated as separate regions (these three regions each contained one gap longer than 130 kb).
Tag SNP portability
PVT, the proportion of variation tagged by tag SNPs, is the fraction of polymorphic non-tag SNPs in a target population that are in LD with at least one tag SNP, above a specified r2 cutoff (Conrad et al., 2006). Our evaluation of PVT followed that of Conrad et al. (2006), with two main modifications (that had very minor effects on the magnitude of PVT). First, a strict MAF cutoff of >0.05 based on the estimated allele frequency of a given SNP in a population was used in place of a chromosome number cutoff method used by Conrad et al. (2006), where the product of the number of chromosomes in the population and 0.05 was rounded to the nearest integer and SNPs with minor allele present on greater than this number of chromosomes were retained for analysis. Second, the tag SNP in a given LD block was chosen to have high r2 values with other SNPs in the block (see below), in place of the procedure of Conrad et al. (2006) that used the first SNP in the block.
Analysis of tag SNP portability was performed using core SNPs that had MAF>0.05 and that overlapped with the HapMap for 29 regions (X-chromosomal regions 23–26, and regions 30–32 that contained gaps were excluded). Of the 1,309 autosomal core SNPs that overlapped with the HapMap, 154 SNPs (regions 30–32) were excluded from the tag SNP analysis, leaving 1,155 SNPs. The number of core SNPs present in the HapMap ranges from 27 to 58 per region, out of a maximum of 60. Separately for each HapMap population (CEU, CHB+JPT, YRI), after excluding SNPs with MAF≤0.05 in that population, r2 was calculated pairwise for all remaining SNPs in each region. In each HapMap group we selected 333 LD-based tag SNPs with the goal of maximizing the number of SNPs that had r2≥0.85 with at least one tag SNP. This choice of 333 SNPs matches that of Conrad et al. (2006), and it leads to a tag SNP density roughly coincident with panels based on ~500,000 SNPs spread across the human genome.
Our tag SNP selection algorithm was based on a modification of the method of Carlson et al. (2004). For each SNP in a given region, the number of SNPs with which it had r2≥0.85 was calculated. All SNPs not in any pairs with r2≥0.85 in the donor population (“singletons”) were excluded from consideration. The SNP(s) that had r2≥0.85 with the largest number of SNPs in the region were then identified. To break ties, all pairwise r2 values above 0.85 that involved at least one of the tied SNPs were ranked, with larger values given higher ranks, between 1 and the total number of values considered. The SNP with the largest rank sum across pairs that contained it was chosen as the tag SNP. In case of a further tie in rank sum, the first SNP in the region among those tied with the largest rank sum was chosen as the tag SNP. For subsequent iterations, the tag SNP and the SNPs that it “tagged” were excluded from consideration as tag SNPs, but were still permitted to be considered as tagged SNPs. For each genomic region, this process – ranking SNPs by the number of pairs with r2≥0.85, breaking ties in this quantity using r2 rank sums, and breaking ties in rank sum by SNP position – was repeated until all SNPs in the region that had r2≥0.85 with at least one other SNP were either chosen as tag SNPs, or were tagged by tag SNPs. If the number of tag SNPs chosen in a population was less than 333, then the tag SNPs were supplemented using singletons randomly chosen from all regions to produce a tag panel containing 333 SNPs. As singletons each tag only one SNP in the donor population – but may tag different numbers of SNPs in target populations – different singleton sets may lead to slightly different values of PVT. Note that no guarantee was made that all genomic regions would contain at least one tag SNP. However, for each HapMap sample, in the tag panel based on that sample, each region did contain at least one tag SNP.
We focused on common variants in our use of the PVT score to measure the amount of variation indirectly assayed in the “target” population by typing markers selected in the “donor” population. In counting polymorphic SNPs among core SNPs in region i (pi, following Conrad et al. (2006), except with regions indexed by i instead of r), we excluded from consideration SNPs that had MAF≤0.05 in the target population. We also excluded SNPs that had MAF≤0.05 in the target population when counting tag SNPs from the donor group (si). Excluding SNPs with MAF≤0.05 in the target population once more, we then determined the number of non-tag core SNPs in the target population that were “tagged” by the tag SNPs from the donor population, or ti-si (ti is the number of polymorphic tagged SNPs, including tag SNPs). To be considered “tagged,” we required that a non-tag SNP have r2≥0.85 with at least one tag SNP. Summing ti-si across regions, we obtained the total number of polymorphic non-tag SNPs in the target population that were tagged by tag SNPs from the donor population. We computed PVT as the ratio of this quantity and the total number of non-tag core SNPs with MAF>0.05 in the target population (that is, pi-si summed across regions). Because sample sizes vary across populations and a linear relationship between PVT and sample size (in the relevant range) has been observed previously (Conrad et al., 2006), PVT scores were adjusted to the mean sample size across HGDP-CEPH populations (36 chromosomes). For populations with more than 36 chromosomes, we adjusted PVT empirically by resampling 36 chromosomes from the population 30 times, averaging PVT across these subsamples. For populations with fewer than 36 chromosomes, we used a regression adjustment to “bring them up” to 36 (Conrad et al., 2006). In cases where this adjustment produced PVT scores above 1, PVT was set to 1.
Tag SNP portability using HapMap mixtures
To examine the ability of tag SNPs selected from mixtures of HapMap samples to capture variation in non-HapMap target populations, various combinations of the HapMap samples were constructed in 5% increments using random subsets of chromosomes. Each subset contained 120 chromosomes, so that a 5% increment corresponded to 6 chromosomes. Subsets were chosen from the 120 chromosomes in CEU and the 120 chromosomes in YRI (excluding offspring in trios), and the 180 chromosomes in CHB+JPT. For each of the 231 combinations of proportions possible using three groups and increments of 5%, r2 was calculated on 30 random subsets of the 420 HapMap chromosomes that represented the HapMap groups in the specified proportions. In the cases of mixture proportions with 100% CEU or YRI representation, the 30 subsets were identical, containing all 120 CEU or YRI chromosomes. For all combinations of proportions, the same HapMap subsets were used for each target population.
For each set of proportions, to allow for the exclusion of SNPs that had MAF≤0.05, SNPs with MAF≤0.05 in at least one of the 30 replicates were excluded from consideration as tag SNPs. After this exclusion, the average r2 values across the 30 replicates were used for the selection of a tag SNP panel comprised of 333 tag SNPs (based on the modified version of the Carlson et al. (2004) algorithm described above). For each tag SNP panel, PVT was calculated for each target population as described above. A similar approach had been applied by Conrad et al. (2006) in the special cases of equal mixtures of two or three HapMap samples. The 231 pairs of PVT values obtained in the Bengalis and Tamilians across all donor mixtures were compared using a two-sided Wilcoxon signed-rank test.
Note that in the mixture analysis (Figure 3B, Figure 4, and Figure 5), all combinations examined are based on samples of size 120 chromosomes, whereas in the analysis of HapMap samples individually (Figure 3A), 180 chromosomes were used in the CHB+JPT group. For an actual association study in a population best tagged with a panel designed from the CHB+JPT group, use of all 180 chromosomes is preferable, but we chose to use 120 in the mixture analysis to achieve a fair comparison. Measurements of PVT increase for optimal mixtures (Figure 3B) are reported relative to the highest-scoring vertex in Figure 4 and Figure 5, representing the highest-scoring individual HapMap sample. Small length differences between gray bars in Figure 3B and corresponding colored bars in Figure 3A are explained partly by the difference in the sets of 120 chromosomes used for Figure 3B from the full CHB+JPT data used for Figure 3A; differences in the choice of singletons in the analyses that underlie the two figures also make a small contribution.
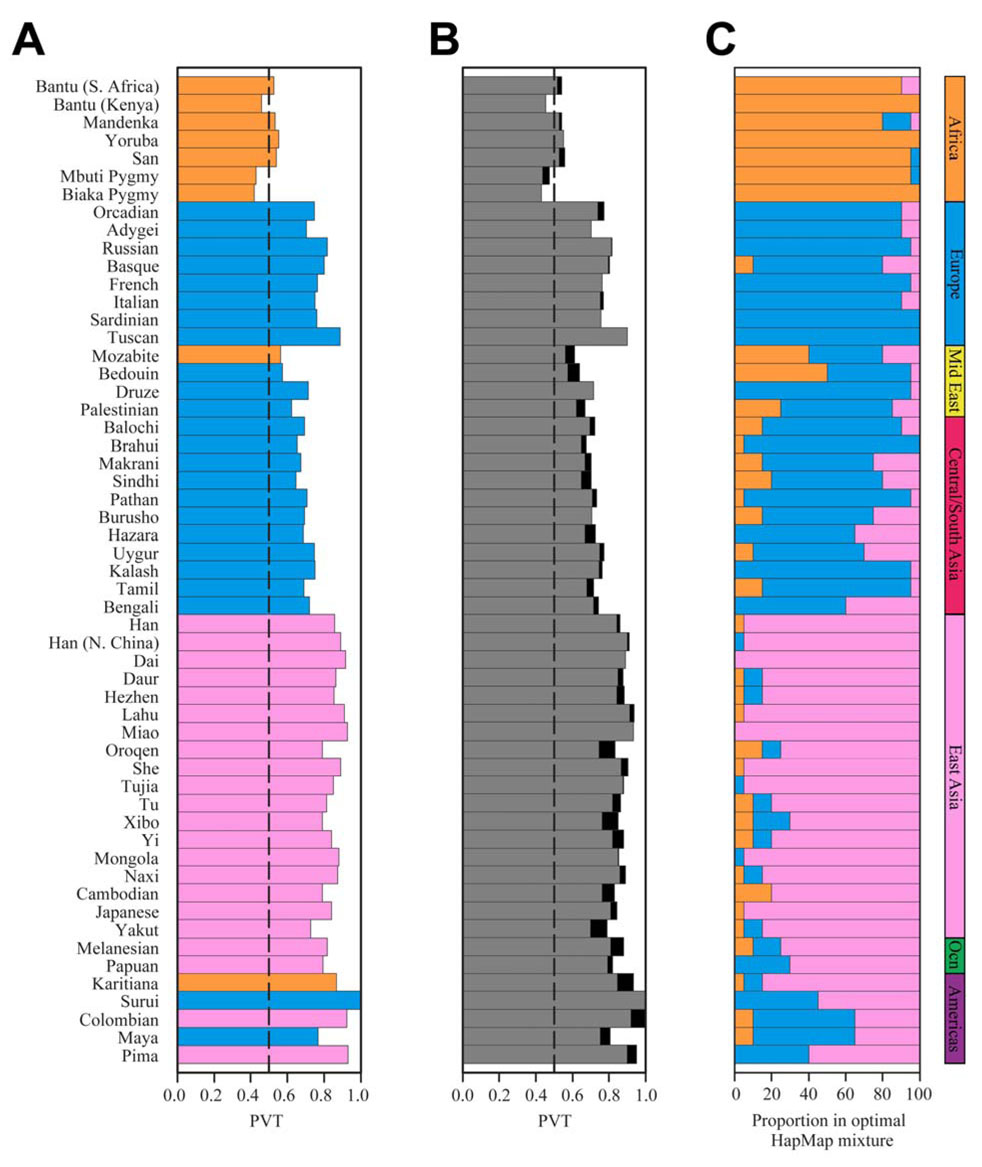
Figure 3.
Portability of tag SNPs chosen using the individual HapMap populations and optimal HapMap mixtures, for each of the 55 populations (as measured by PVT). (A) The proportion of polymorphic non-tag SNPs with MAF>0.05 in the target population that have r2≥0.85 with at least one tag SNP (PVT). PVT is plotted only for the HapMap group that produced the highest PVT. For each population, the color of the bar indicates the HapMap sample from which the optimal tag SNP set was chosen (blue=CEU, pink=CHB+JPT, orange=YRI). The vertical line indicates 50% tag portability. (B) The highest PVT obtained using tag SNP panels from HapMap mixtures. The black portion of the bar represents the increase in PVT obtained using tag SNPs from the optimal HapMap mixture compared to the most effective individual HapMap sample. (C) The proportions of the three HapMap populations in the optimal HapMap mixture that produced the highest PVT (blue=CEU, pink=CHB+JPT, orange=YRI). In the Surui and Colombian populations, multiple mixtures produced PVT values above 1, and the optimal mixture was chosen as the one with the highest unadjusted PVT (the same procedure was applied for Surui in part A).
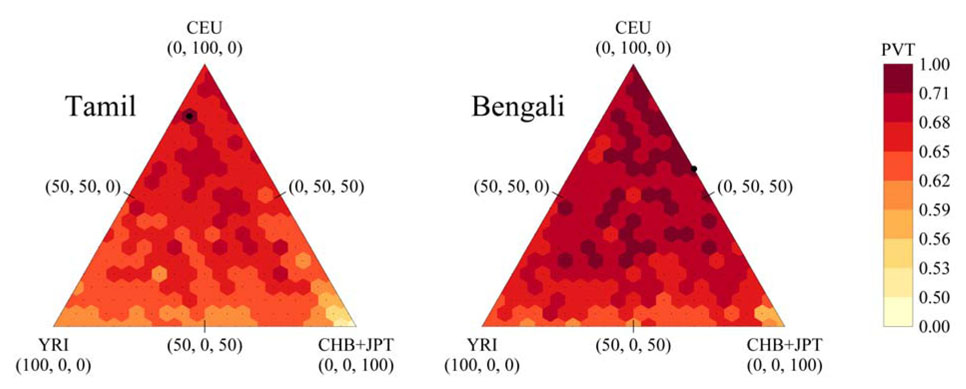
Figure 4.
Portability in the Tamilians and Bengalis of tag SNPs chosen from different mixtures of HapMap populations, as measured by PVT. Each vertex of the triangle represents one of the three HapMap populations (CEU, CHB+JPT, YRI), with increasing distance from that vertex indicating a smaller percentage of that HapMap population present in the population mixture. The shading represents the level of portability as measured by PVT. Note that the darkest and lightest colors represent wider ranges of PVT values than the other colors. A black circle indicates the combination of the three HapMap samples that produces the highest PVT among the points tested (80% CEU, 5% CHB+JPT, 15% YRI for Tamilians; 60% CEU, 40% CHB+JPT, 0% YRI for Bengalis).
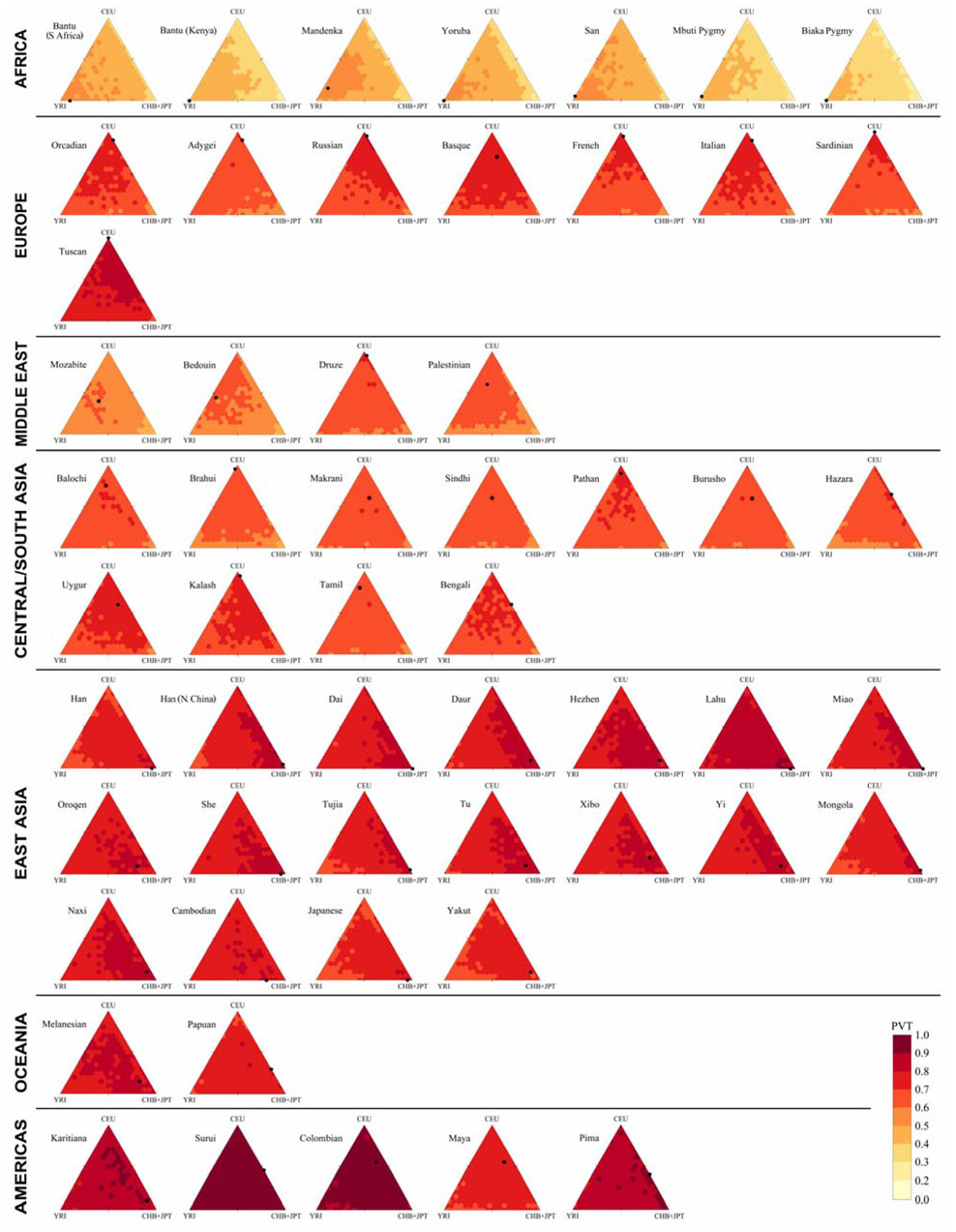
Figure 5.
Portability in individual populations of tag SNPs chosen from different mixtures of HapMap populations, as measured by PVT. The figure design follows that of Figure 4, with a different color scale.
FST to the nearest HapMap population
FST was evaluated based on the same 1,155 SNPs as those used in the tag SNP analysis. Eq. 5.3 of Weir (1996) was applied to each genomic region. After setting negative values to zero, estimates were averaged across regions to obtain the overall estimate.
Results
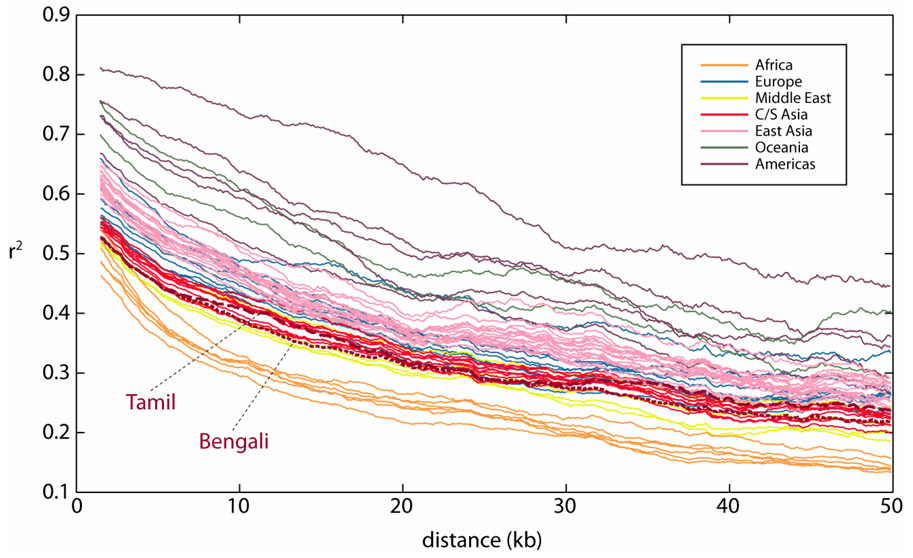
Figure 1 shows the decay of LD in the various populations, illustrating that the level of LD in the Indian populations is relatively low in comparison with that in other non-African groups. Sub-Saharan African populations have the lowest level of LD, followed by populations from the Middle East (including North Africa), Central/South Asia, Europe, East Asia, Oceania, and the Americas. Averaging across populations within geographic regions, LD levels drop below r2=0.5 at 1.4 kb for Africa, 2.6 kb for the Middle East, 3.3 kb for Central/South Asia, 6.1 kb for Europe, 9.8 kb for East Asia, 15.7 kb for Oceania, and 21.6 kb for the Americas. LD in Bengalis and Tamilians is similar to that in other Central/South Asian populations. Averaging the values for the two Indian groups, LD reaches r2<0.5 at 2.9 kb.
Figure 1.
Linkage disequilibrium vs. physical distance. The r2 statistic was calculated for each pair of SNPs with MAF≥0.1. The mean r2 for a given distance bin is plotted as a function of the mean distance between pairs of SNPs with distance in the bin. Bin size is 6 kb. Each line represents a separate population.
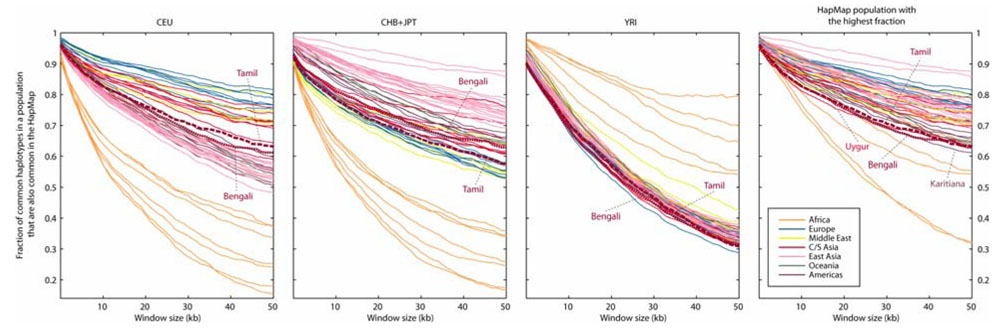
As measured by haplotype sharing, the HapMap captures common haplotypes relatively well in most HGDP-CEPH populations (Conrad et al., 2006). When we include the Bengalis and Tamilians, we notice that among the non-African populations, the fraction of common haplotypes also common in the most similar HapMap population is lowest in Bengalis and Tamilians, as well as in the Uygur and Karitiana populations – of western China and the Amazon region, respectively (Figure 2). The CEU group captures common haplotypes in the Tamilians to a greater extent than does the CHB+JPT group, and CHB+JPT captures common haplotypes in the Bengalis to a greater extent than does CEU. This result is compatible with the proximity to East Asia of the Bengalis in northeast India, in comparison with the greater distance to East Asia for the Tamilians in southern India, and with the similarity to East Asians detected in Bengalis in analysis of unlinked markers from the same individuals (Rosenberg et al., 2006).
Figure 2.
The fraction of common haplotypes (≥10% frequency) in individual populations that are also common in the HapMap. For each plot we used haplotypes based on the SNPs that overlap between HapMap Phase II and our autosomal core regions, and we averaged over all windows of a given length. The graph on the right shows the fraction of the common haplotypes of a population that are also common in the most similar HapMap sample (determined point by point). Thus, for each population and each window size, the rightmost panel takes the highest value among those shown in the other three panels. The non-African populations with the lowest level of coverage by the most similar HapMap population are labeled in the rightmost panel.
Considering each of the three HapMap populations as donor populations for selection of tag SNPs, variation in the Indian populations is tagged most effectively by CEU (Figure 3A). Among non-African populations worldwide, the Bengalis and Tamilians are the 12th and 6th least effectively tagged. However, when we compare a tag SNP set based on the optimal HapMap mixture to the tag SNP set based on CEU, PVT increases by 5.1% in Tamilians and by 4.1% in Bengalis. Using optimal HapMap mixtures, the proportion of variation tagged increases by larger amounts in many other populations (Figure 3B) – the relative increases in Tamilians and Bengalis were the 19th and 27th largest among all 55 populations. The greatest increases were 12.8%, 11.8%, 11.8% and 11.3% in Yakut, Oroqen, Xibo, and Bedouin, respectively, and the average gain was 4.2%. Populations from Africa and Europe showed relatively little change with optimal HapMap mixtures compared to using the individual HapMap sample that produced the highest PVT (average of 3.0% for populations from Africa and 1.0% for Europe). Populations from East Asia (4.7%) and from geographic regions more distant from the HapMap samples – Central/South Asia (4.3%), the Middle East (7.0%), Oceania (6.0%), and the Americas (6.3%) – had somewhat greater increases. The Spearman correlation of the percent gain in PVT with the FST genetic distance to the most genetically similar HapMap population equals 0.392 (P=0.003), indicating that the degree to which the mixture method increases the proportion of variation tagged in a population is correlated with the genetic proximity of the population to one of the HapMap populations. In East Asia, the only geographic region represented by the HapMap in which PVT gains were substantial, the largest increases were observed in the relatively divergent Yakut, Oroqen, and Xibo populations.
The full results of the tag SNP analysis with HapMap mixtures are shown in Figure 4 and Figure 5 as equilateral triangles in which the vertices represent PVT for tag panels based only on CEU, CHB+JPT, or YRI, and in which interior points show PVT values for appropriate mixtures. The Bengalis had higher PVT than the Tamilians for nearly all donor mixtures (229 of 231, P<0.001). Both groups showed reduced PVT near the CHB+JPT and YRI vertices, and increased PVT near the CEU vertex (Figure 4). The Bengalis were optimally tagged by a combination (60% CEU, 40% CHB+JPT, 0% YRI) similar to the optimal combination of Europeans and East Asians for predicting allele frequencies in India (Rosenberg et al., 2006). The optimal donor mixture for Tamilians also had majority representation from CEU (80%); however, the remaining 20% was split between YRI (15%) and CHB+JPT (5%).
With five exceptions, the major contributing HapMap sample in the mixture that provided the optimal tag SNP panel was the same group that best captured variation in the population when HapMap samples were evaluated separately (Figure 3C and Figure 5). In Karitiana, YRI produced the highest PVT among the three HapMap samples (0.869), slightly higher than for CHB+JPT (0.865); however, the largest fraction in the optimal mixture was from CHB+JPT, with YRI present at only 5%. In Surui, CEU produced the highest PVT individually, while CHB+JPT had the largest share in the optimal mixture; the reverse was true for Colombians. In Bedouins, CEU produced the highest PVT, but the largest fraction in the optimal mixture was from YRI; in Mozabites, YRI produced the highest PVT, and the optimal mixture had equal CEU and YRI components. Although these exceptions were unusual, optimal mixtures for some populations in the Americas, Central/South Asia, and the Middle East contained sizeable proportions of a different HapMap sample from the one that produced the highest PVT individually.
Discussion
Linkage disequilibrium in Indian populations has generally been investigated only for smaller numbers of SNPs (Tang et al., 2002, Vishwanathan et al., 2003, Cha et al., 2004, Sengupta et al., 2004, Beaty et al., 2005, Raj et al., 2006, Prasad & Thelma, 2007, Roy et al., 2008), and has not been extensively compared with LD in other populations. We found that the Bengalis and Tamilians have a similar level of LD to other populations in the surrounding geographical area – but lower LD than in Europe or East Asia. Haplotype variation in the Bengalis and Tamilians is relatively poorly captured by the HapMap, when using HapMap samples individually. However, when employing combinations of the three HapMap samples in tag SNP selection, the proportion of variation tagged increased in the Bengalis and Tamilians by a modest but noticeable 5.1% and 4.1%, respectively, and a gain of up to ~12% was achieved for other populations. The degree to which this mixture method increases the proportion of variation tagged in a population is associated with the genetic proximity of the population to one of the HapMap populations, with the largest increases being observed in geographic regions distant from the HapMap populations.
The mixture approach we have discussed here can be considered as a complementary tag SNP selection strategy to methods that identify tag SNP panels applicable to multiple populations (Ahmadi et al., 2005, Howie et al., 2006, Xu et al., 2007a, 2007b). Such methods produce tag SNP sets that are not likely to be optimal in any particular population, but that are generally useful across a wide range of populations. By contrast, the mixture method takes the approach of producing more specifically customized optimal panels for individual populations, and is likely to be of greatest use when a study is planned for one or a small number of closely related non-HapMap groups. In such cases, before a full-scale tag SNP association study is performed, some level of preliminary SNP genotype data – preferably chosen to be representative according to genomic variables such as recombination rate, gene density, and sequence conservation – is required from the non-HapMap population of interest, so that the ideal mixture for use in the population can be evaluated. Thus, a limitation of our mixture method is that its utility is restricted to situations for which such initial data are feasible to obtain.
It is noteworthy that our method of choosing tag SNPs in sample mixtures relies on r2 computations in structured populations, so that some SNP pairs may have had their LD estimates inflated by population structure (Nei & Li, 1973, Ohta, 1982). However, at short distances the effect of population structure on LD is likely to be relatively small, as suggested by the fact that the local decay of LD is quite similar in West Africans and closely related African Americans who have European admixture (Gabriel et al., 2002). Because PVT in target populations increased when using tag SNPs obtained from donor mixtures, it is likely that at short distances, any effect of population structure on r2 is outweighed by the increase in tagging potential produced when considering more than one HapMap sample in the selection of tag SNPs. Although in our study, the experimental design using discrete genomic regions protects against the possibility of long-range correlations induced by population structure, in applications of the mixture approach on a full genomic scale it may be advisable to limit the distance allowed between tag SNPs and tagged SNPs.
The observation that PVT was higher for Bengalis than for Tamilians likely results from greater similarity of Bengalis to the relatively well-tagged populations of East Asia. The optimal combination of the individual HapMap samples for tag SNP selection differed between the Bengalis and Tamilians, having a greater contribution from CHB+JPT in Bengalis. This greater proportion from CHB+JPT for Bengalis could reflect a greater degree of East Asian gene flow into northeast India, with the effects of this gene flow not having reached as far as southern India. Perhaps due to small sample sizes that may have produced somewhat imprecise r2 estimates and flat PVT surfaces as a function of the mixture coefficients, some uncertainty was visible in choosing the optimal mixture, as multiple mixtures often produced similar PVT values close to the maximum (Figure 4 and Figure 5); the precise location of the maximum may also fluctuate with the portions of the genome studied. In general, however, the major contributing HapMap sample in the mixture that provided the optimal tag SNP panel (Figure 3C) was the same group that best captured variation in the population when evaluating HapMap samples separately (Figure 3A). In addition, especially for some populations in the Americas, Central/South Asia, and the Middle East, optimal mixtures contained sizeable components from more than one HapMap sample.
As can be observed from a comparison of Figures 3A and 3B, the rank ordering of populations by PVT values does not differ dramatically when using optimal mixtures compared to using individual HapMap samples (Spearman’s ρ=0.990). Thus, while some increase in tagging potential is observed in optimal mixtures, especially in Asian populations not closely related to the HapMap samples, the identities of the populations most difficult to tag are not substantially changed by the use of mixtures. While the PVT rank order will change as large-scale studies expand to incorporate new populations, our mixture-based approach is still likely to provide a way of extracting additional tagging information in the populations left by next-generation databases with the smallest level of genomic coverage.
Finally, the mixture strategy we propose, which we have applied to the tag SNP selection problem, can be viewed as a general approach for applying genomic databases built in small numbers of populations for use in a wider variety of groups. A related situation occurs when HapMap data are used for imputing missing genotypes in non-HapMap populations to facilitate the testing of untyped SNPs for genetic association with phenotypes (Marchini et al., 2007, Servin & Stephens, 2007). In that context, the use in non-HapMap populations of mixtures of HapMap datasets may have the potential to improve the imputation of missing genotypes and thereby to increase the power of subsequent association tests.
Acknowledgments
We thank J. DeYoung and the Southern California Genotyping Consortium for genotyping. The unpublished genotypes of 160 African individuals used during the data cleaning phase of this study were obtained in collaboration with F. Reed and S. Tishkoff. This investigation was supported by a pilot grant award from the Center of Excellence in Genomic Science at the University of Southern California (T.J.P.), a University of Michigan Center for Genetics in Health and Medicine postdoctoral fellowship (M.J.), NIH grant GM081441 (N.A.R.), and grants from the Burroughs Wellcome Fund (J.K.P., N.A.R.), the Alfred P. Sloan Foundation (J.D.W., J.K.P., N.A.R.), the Packard Foundation (J.K.P.), and the National Science Foundation (J.D.W.). The research was conducted in part in a facility constructed with support from Research Facilities Improvement Program grant C06 (RR10600-01, CA62528-01, RR14514-01) from the National Center for Research Resources, National Institutes of Health.
Abbreviations
- CEPH
Centre d’Etude du Polymorphisme Humain
- HGDP
Human Genome Diversity Project
- LD
linkage disequilibrium
- MAF
minor allele frequency
- PVT
proportion of variation tagged
- SNP
single nucleotide polymorphism
Footnotes
Publisher's Disclaimer: This PDF receipt will only be used as the basis for generating PubMed Central (PMC) documents. PMC documents will be made available for review after conversion (approx. 2–3 weeks time). Any corrections that need to be made will be done at that time. No materials will be released to PMC without the approval of an author. Only the PMC documents will appear on PubMed Central -- this PDF Receipt will not appear on PubMed Central.
References
- Ahmadi KR, Weale ME, Xue ZY, Soranzo N, Yarnall DP, Briley JD, Maruyama Y, Kobayashi M, Wood NW, Spurr NK, Burns DK, Roses AD, Saunders AM, Goldstein DB. A single-nucleotide polymorphism tagging set for human drug metabolism and transport. Nat Genet. 2005;37:84–89. doi: 10.1038/ng1488. [DOI] [PubMed] [Google Scholar]
- Andrés AM, Clark AG, Shimmin L, Boerwinkle E, Sing CF, Hixson JE. Understanding the accuracy of statistical haplotype inference with sequence data of known phase. Genet Epidemiol. 2007;31:659–671. doi: 10.1002/gepi.20185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Fallin MD, Hetmanski JB, McIntosh I, Chong SS, Ingersoll R, Sheng X, Chakraborty R, Scott AF. Haplotype diversity in 11 candidate genes across four populations. Genetics. 2005;171:259–267. doi: 10.1534/genetics.105.043075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand M-F, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, Chen Z, Chu J, Carcassi C, Contu L, Du R, Excoffier L, Ferrara GB, Friedlaender JS, Groot H, Gurwitz D, Jenkins T, Herrera RJ, Huang X, Kidd J, Kidd KK, Langaney A, Lin AA, Mehdi SQ, Parham P, Piazza A, Pistillo MP, Qian Y, Shu Q, Xu J, Zhu S, Weber JL, Greely HT, Feldman MW, Thomas G, Dausset J, Cavalli-Sforza LL. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha P-C, Yamada R, Sekine A, Nakamura Y, Koh C-L. Inference from the relationships between linkage disequilibrium and allele frequency distributions of 240 candidate SNPs in 109 drug-related genes in four Asian populations. J Hum Genet. 2004;49:558–572. doi: 10.1007/s10038-004-0190-z. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- De Bakker PI, Graham RR, Altshuler D, Henderson BE, Haiman CA. Transferability of tag SNPs to capture common genetic variation in DNA repair genes across multiple populations. Pac Symp Biocomput. 2006a;11:478–486. [PubMed] [Google Scholar]
- De Bakker PIW, Burtt NP, Graham RR, Guiducci C, Yelensky R, Drake JA, Bersaglieri T, Penney KL, Butler J, Young S, Onofrio RC, Lyon HN, Stram DO, Haiman CA, Freedman ML, Zhu X, Cooper R, Groop L, Kolonel LN, Henderson BE, Daly MJ, Hirschhorn JN, Altshuler D. Transferability of tag SNPs in genetic association studies in multiple populations. Nat Genet. 2006b;38:1298–1303. doi: 10.1038/ng1899. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- González-Neira A, Ke X, Lao O, Calafell F, Navarro A, Comas D, Cann H, Bumpstead S, Ghori J, Hunt S, Deloukas P, Dunham I, Cardon LR, Bertranpetit J. The portability of tagSNPs across populations: a worldwide survey. Genome Res. 2006;16:323–330. doi: 10.1101/gr.4138406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Pakstis AJ, Li H, Speed WC, Kidd JR, Kidd KK. Significant variation in haplotype block structure but conservation in tagSNP patterns among global populations. Eur J Hum Genet. 2007;15:302–312. doi: 10.1038/sj.ejhg.5201751. [DOI] [PubMed] [Google Scholar]
- Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- Howie BN, Carlson CS, Rieder MJ, Nickerson DA. Efficient selection of tagging single-nucleotide polymorphisms in multiple populations. Hum Genet. 2006;120:58–68. doi: 10.1007/s00439-006-0182-5. [DOI] [PubMed] [Google Scholar]
- Huang W, He Y, Wang H, Wang Y, Liu Y, Wang Y, Chu X, Wang Y, Xu L, Shen Y, Xiong X, Li H, Wen B, Qian J, Yuan W, Zhang C, Wang Y, Jiang H, Zhao G, Chen Z, Jin L. Linkage disequilibrium sharing and haplotype-tagged SNP portability between populations. Proc Nat Acad Sci USA. 2006;103:1418–1421. doi: 10.1073/pnas.0510360103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A, Vavruch-Nilsson V, Cox DR, Frazer KA, Gyllensten U. Evaluation of the SNP tagging approach in an independent population sample: array-based SNP discovery in Sami. Hum Genet. 2007;122:141–150. doi: 10.1007/s00439-007-0379-2. [DOI] [PubMed] [Google Scholar]
- Landwehr N, Mielikäinen T, Eronen L, Toivonen H, Mannila H. Constrained hidden Markov models for population-based haplotyping. BMC Bioinformatics. 2007;8:S9. doi: 10.1186/1471-2105-8-S2-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li J. Comparison of haplotyping methods using families and unrelated individuals on simulated rheumatoid arthritis data. BMC Proc. 2007;1:S55. doi: 10.1186/1753-6561-1-s1-s55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Kim YJ, Yoon Y, Kim SO, Kang H, Park J, Han AR, Han B, Oh B, Kimm K, Yoon B, Song K. Comparative study of the linkage disequilibrium of an ENCODE region, chromosome 7p15, in Korean, Japanese, and Han Chinese samples. Genomics. 2006;87:392–398. doi: 10.1016/j.ygeno.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Mahasirimongkol S, Chantratita W, Promso S, Pasomsab E, Jinawath N, Jongjaroenprasert W, Lulitanond V, Krittayapoositpot P, Tongsima S, Sawanpanyalert P, Kamatani N, Nakamura Y, Sura T. Similarity of the allele frequency and linkage disequilibrium pattern of single nucleotide polymorphisms in drug-related gene loci between Thai and northern East Asian populations: implications for tagging SNP selection in Thais. J Hum Genet. 2006;51:896–904. doi: 10.1007/s10038-006-0041-1. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Marvelle AF, Lange LA, Qin L, Wang Y, Lange EM, Adair LS, Mohlke KL. Comparison of ENCODE region SNPs between Cebu Filipino and Asian HapMap samples. J Hum Genet. 2007;52:729–737. doi: 10.1007/s10038-007-0175-9. [DOI] [PubMed] [Google Scholar]
- Montpetit A, Nelis M, Laflamme P, Magi R, Ke X, Remm M, Cardon L, Hudson TJ, Metspalu A. An evaluation of the performance of tag SNPs derived from HapMap in a Caucasian population. PLoS Genet. 2006;2:282–290. doi: 10.1371/journal.pgen.0020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JC, Lõhmussaar E, Mägi R, Remm M, Bettecken T, Lichtner P, Biskup S, Illig T, Pfeufer A, Luedemann J, Schreiber S, Pramstaller P, Pichler I, Romeo G, Gaddi A, Testa A, Wichmann H-E, Metspalu A, Meitinger T. Linkage disequilibrium patterns and tagSNP transferability among European populations. Am J Hum Genet. 2005;76:387–398. doi: 10.1086/427925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Li W-H. Linkage disequilibrium in subdivided populations. Genetics. 1973;75:213–219. doi: 10.1093/genetics/75.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. Linkage disequilibrium due to random genetic drift in finite subdivided populations. Proc Nat Acad Sci USA. 1982;79:1940–1944. doi: 10.1073/pnas.79.6.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad P, Thelma BK. Normative genetic profiles of RAAS pathway gene polymorphisms in North Indian and South Indian populations. Hum Biol. 2007;79:241–254. doi: 10.1353/hub.2007.0033. [DOI] [PubMed] [Google Scholar]
- Raj SM, Chakraborty R, Wang N, Govindaraju DR. Linkage disequilibria and haplotype structure of four SNPs of the interleukin 1 gene cluster in seven Asian Indian populations. Hum Biol. 2006;78:109–119. doi: 10.1353/hub.2006.0026. [DOI] [PubMed] [Google Scholar]
- Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, Lander ES. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- Ribas G, González-Neira A, Salas A, Milne RL, Vega A, Carracedo B, González E, Barroso E, Fernández LP, Yankilevich P, Robledo M, Carracedo A, Benítez J. Evaluating HapMap SNP data transferability in a large-scale genotyping project involving 175 cancer-associated genes. Hum Genet. 2006;118:669–679. doi: 10.1007/s00439-005-0094-9. [DOI] [PubMed] [Google Scholar]
- Roberts A, McMillan L, Wang W, Parker J, Rusyn I, Threadgill D. Inferring missing genotypes in large SNP panels using fast nearest-neighbor searches over sliding windows. Bioinformatics. 2007;23:i401–i407. doi: 10.1093/bioinformatics/btm220. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Mahajan S, Gonzalez-Quevedo C, Blum MGB, Nino-Rosales L, Ninis V, Das P, Hegde M, Molinari L, Zapata G, Weber JL, Belmont JW, Patel PI. Low levels of genetic divergence across geographically and linguistically diverse populations of India. PLoS Genet. 2006;2:2052–2061. doi: 10.1371/journal.pgen.0020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy NS, Farheen S, Roy N, Sengupta S, Majumder PP. Portability of tag SNPs across isolated population groups: an example from India. Ann Hum Genet. 2008;72:82–89. doi: 10.1111/j.1469-1809.2006.00383.x. [DOI] [PubMed] [Google Scholar]
- Sawyer SL, Mukherjee N, Pakstis AJ, Feuk L, Kidd JR, Brookes AJ, Kidd KK. Linkage disequilibrium patterns vary substantially among populations. Eur J Hum Genet. 2005;13:677–686. doi: 10.1038/sj.ejhg.5201368. [DOI] [PubMed] [Google Scholar]
- Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Farheen S, Mukherjee N, Dey B, Mukhopadhyay B, Sil SK, Prabhakaran N, Ramesh A, Edwin D, Usha Rani MV, Mitra M, Mahadik CT, Singh S, Sehgal SC, Majumder PP. DNA sequence variation and haplotype structure of the ICAM1 and TNF genes in 12 ethnic groups of India reveal patterns of importance in designing association studies. Ann Hum Genet. 2004;68:574–587. doi: 10.1046/j.1529-8817.2003.00126.x. [DOI] [PubMed] [Google Scholar]
- The International Collaborative Group on Isolated Populations. Service S, Sabatti C, Freimer N. Tag SNPs chosen from HapMap perform well in several population isolates. Genet Epidemiol. 2007;31:189–194. doi: 10.1002/gepi.20201. [DOI] [PubMed] [Google Scholar]
- Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:1296–1308. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EM, Wang X, Littrell J, Eckert J, Cole R, Kissebah AH, Olivier M. Comparison of linkage disequilibrium patterns between the HapMap CEPH samples and a family-based cohort of Northern European descent. Genomics. 2006;88:407–414. doi: 10.1016/j.ygeno.2006.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankovich J, Cox CJ, Tan RB, Montgomery DS, Huxtable SJ, Rubio JP, Ehm MG, Johnson L, Butzkueven H, Kilpatrick TJ, Speed TP, Roses AD, Bahlo M, Foote SJ. On the utility of data from the International HapMap Project for Australian association studies. Hum Genet. 2006;119:220–222. doi: 10.1007/s00439-005-0120-y. [DOI] [PubMed] [Google Scholar]
- Tang K, Ngoi S-M, Gwee P-C, Chua JMZ, Lee EJD, Chong SS, Lee CGL. Distinct haplotype profiles and strong linkage disequilibrium at the MDR1 multidrug transporter gene locus in three ethnic Asian populations. Pharmacogenetics. 2002;12:437–450. doi: 10.1097/00008571-200208000-00004. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Kidd KK. Implications of biogeography of human populations for 'race' and medicine. Nat Genet. 2004;36:S21–S27. doi: 10.1038/ng1438. [DOI] [PubMed] [Google Scholar]
- Vishwanathan H, Edwin D, Usha Rani MV, Majumder PP. A survey of haplotype frequencies and linkage disequilibrium at the DRD2 locus in the Nilgiri hill tribes, South India. Curr Sci. 2003;84:566–570. [Google Scholar]
- Weir BS. Genetic Data Analysis II. Sunderland, MA: Sinauer Associates; 1996. [Google Scholar]
- Willer CJ, Scott LJ, Bonnycastle LL, Jackson AU, Chines P, Pruim R, Bark CW, Tsai Y-Y, Pugh EW, Doheny KF, Kinnunen L, Mohlke KL, Valle TT, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. Tag SNP selection for Finnish individuals based on the CEPH Utah HapMap database. Genet Epidemiol. 2006;30:180–190. doi: 10.1002/gepi.20131. [DOI] [PubMed] [Google Scholar]
- Xu Z, Kaplan NL, Taylor JA. Tag SNP selection for candidate gene association studies using HapMap and gene resequencing data. Eur J Hum Genet. 2007a;15:1063–1070. doi: 10.1038/sj.ejhg.5201875. [DOI] [PubMed] [Google Scholar]
- Xu Z, Kaplan NL, Taylor JA. TAGster: efficient selection of LD tag SNPs in single or multiple populations. Bioinformatics. 2007b;23:3254–3255. doi: 10.1093/bioinformatics/btm426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Schaid DJ. Methods to impute missing genotypes for population data. Hum Genet. 2007;122:495–504. doi: 10.1007/s00439-007-0427-y. [DOI] [PubMed] [Google Scholar]