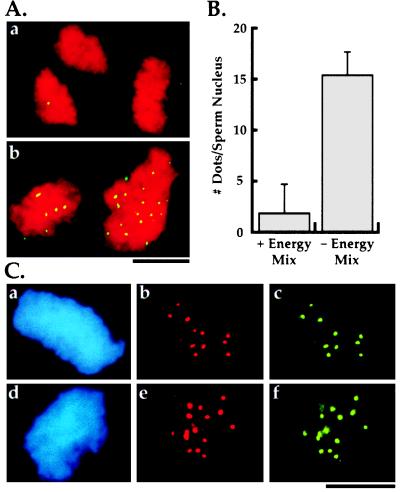
Figure 3.
Characterization of in vitro kinetochore assembly. (A) Effect of presence (a) or absence (b) of energy mix in preincubation buffer on kinetochore assembly. DNA is in red and CENP-E is in green. The same result is obtained with anti-dynein and anti-XKCM1 antibodies. There is no striking difference in the morphology of the condensed DNA at the end of the assembly reaction between the two conditions. (B) Quantitation of kinetochore assembly and effect of energy mix in preincubation buffer. The bars represent the mean number of dots of kinetochore marker proteins per sperm nucleus ± 1 SD. For the left column (+ Energy Mix in preincubation buffer) n = 103 sperm nuclei from two experiments on two different extract preparations. For the right column (− Energy Mix in preincubation buffer; standard kinetochore assembly reaction) n = 303 sperm nuclei from four experiments on three different extract preparations. The absence of energy mix in the preincubation buffer results in >90% of the sperm nuclei demonstrating colocalization of all three markers at the reported density. The residual sperm nuclei show no localization of any of the three markers. (Bars = 10 μm.) (C) Double label immunofluorescence of in vitro assembled kinetochores demonstrating colocalization of all three mitotic markers to discrete foci on the DNA. Panels show 4′,6′diamidino-2-phenylindole (DAPI)-labeled sperm nucleus DNA (a and d) and corresponding double label localization of dynein and CENP-E (b and c) or dynein and XKCM1 (e and f). Dots associated with the chromosome clusters are distributed over several focal planes; the images shown are photomicrographs of single focal planes and do not account for all dots present on these chromosomal clusters.