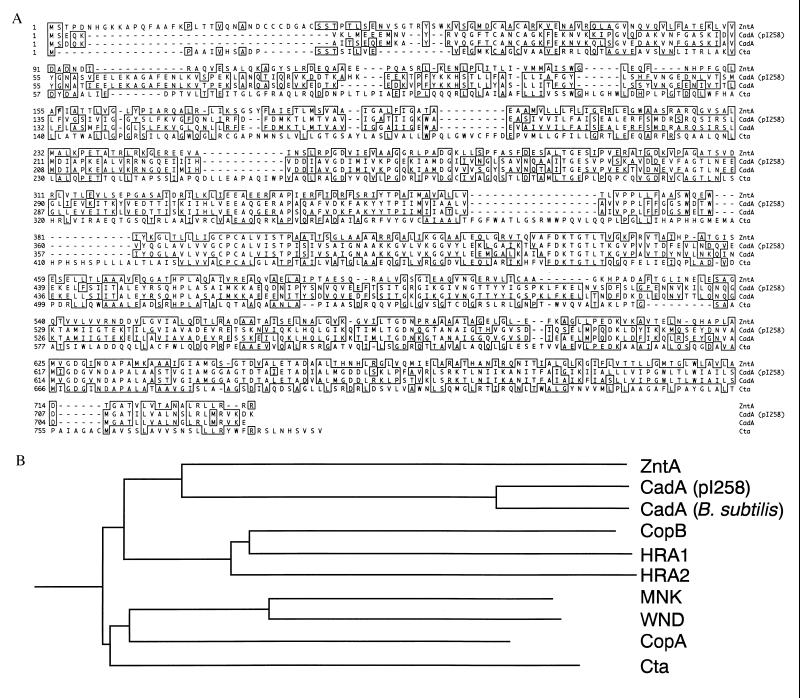
Figure 1.
Multiple alignment and phylogenetic relationship of the amino acid sequences of soft metal translocating P-type ATPases. (A) Alignment of the deduced amino acid sequence of ZntA (22); CadA (pI258) (14); CadA, Bacillus firmus (16); and CtaA, Synechococcus 7942 (18). Identical residues are boxed. (B) The dendogram was made with MegAlign (DNAstar). The branch lengths in the phylogenetic tree are proportional to the number of amino acid substitutions separating each pair. Proteins: ZntA (22), CadA (pI258) (14); CadA, B. firmus (16); CopB, Enterococcus hirae (17); HRA1, E. coli (21); HRA2, E. coli (21); Menkes protein (MNK) (19); Wilson protein (WND) (20); CopA, Enterococcus hirae (17); and CtaA, Synechococcus 7942 (18).