Table 1. Crystal data and data-collection and refinement statistics of the HLA-B*1501 structure.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Wavelength (Å) | 0.907 |
| Unit-cell parameters (Å) | a = 50.66, b = 81.71, c = 109.39 |
| Space group | P212121 |
| Molecules per ASU | 1 |
| Resolution range (Å) | 18.29–1.87 (1.97–1.87) |
| No. of unique reflections | 38220 (5506) |
| Multiplicity | 4.0 (4.1) |
| Completeness of data (%) | 99.8 (100.0) |
| Rmerge† | 0.064 (0.362) |
| 〈I/σ(I)〉 | 15.7 (3.1) |
| Model details | |
| Protein atoms | 3184 |
| Water O atoms | 357 |
| PEG (PG4) atoms | 13 |
| HEPES (EPE) atoms | 15 |
| Refinement details | |
| No. of reflections used in refinement | 36303 |
| R factor for all data‡ (%) | 18.8 |
| Rfree§ (%) | 22.7 |
| Model | |
| Mean isotropic equivalent B factor (Å2) | 25.8 |
| MHC-I heavy-chain B factor (Å2) | 22.3 |
| β2-Microglobulin B factor (Å2) | 31.3 |
| Peptide B factor (Å2) | 21.2 |
| Water B factor (Å2) | 35.1 |
| Heterocompounds B factor (Å2) | 51.0 |
| Residues in most favoured regions¶ (%) | 91.4 |
| Residues in generally disallowed regions¶ (%) | 0.0 |
R
merge = 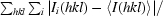
 .
.
R = 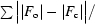
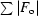 .
.
As R, but calculated on 5% of the data excluded from the refinement.
Calculated with PROCHECK (Laskowski et al., 1993 ▶).
