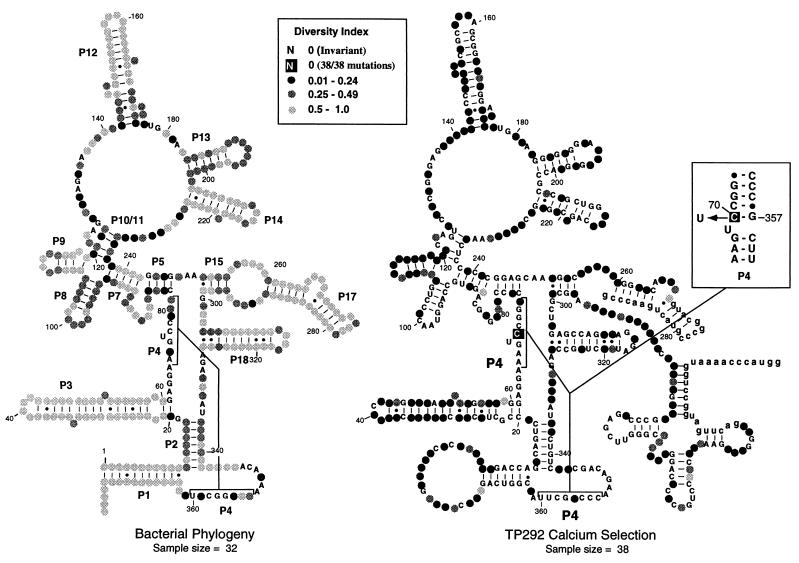
Figure 3.
Distribution of mutations identified in round-18 clones. The genetic variability of each nucleotide position within RNase P/TP292 is summarized for both natural bacterial phylogeny (Left; sequences obtained from the RNase P RNA database; ref. 40) and the Ca2+ selection (Right). The diversity of sequence at each position was calculated as H/Hmax, where H is the Shannon entropy [−Σplog(p)] and Hmax is log(5), for five character states (G, A, U, C, −). Higher sequence variability (i.e., a higher diversity index) is denoted by more lightly shaded circles, and invariant positions are denoted by capital letters. The boxed base at position 70 of TP292 signifies that all round-18 clones contained a mutation at this position. The boxed Inset (Right) shows the position of the C-to-U transition at position 70, within the context of helix P4.