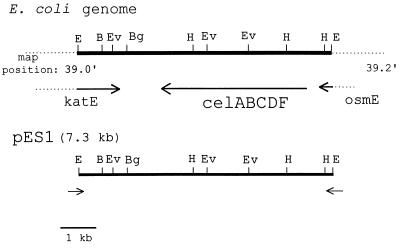
Figure 1.
Comparison of pES1 to sequenced E. coli genomic map position at 39 min. The isolated cosmid clone capable of restoring (GlcNAc)2 utilization in the E. coli transposon mutant strain, Xm1.4, was subcloned to 7.3 kb in the vector Supercos1. Using primers designed to the ends of Supercos1 and reading in the direction of the insert from both directions, we gathered the following sequence data: (i) clockwise (CW), 250 bp; and (ii) counterclockwise (CCW), 225 bp. A blast search of the nucleotide sequence database revealed greater than 99% identity to a region in the osmE gene (CW) and the katE gene (CCW), respectively. Restriction analysis of the clone was in agreement with that reported for the E. coli genome at the region illustrated. Restriction sites shown include B = BamHI, Bg = BglII, E = EcoRI, Ev = EcoRV, and H = HindIII. All other restriction enzymes tested gave results in agreement with the predicted sequence and included the following: AatII, AccI, AvaI, Bsp106, HpaI, KpnI, NdeI, NheI, NotI, PstI, PvuI, SalI, SphI, and XmnI.