Figure 1.
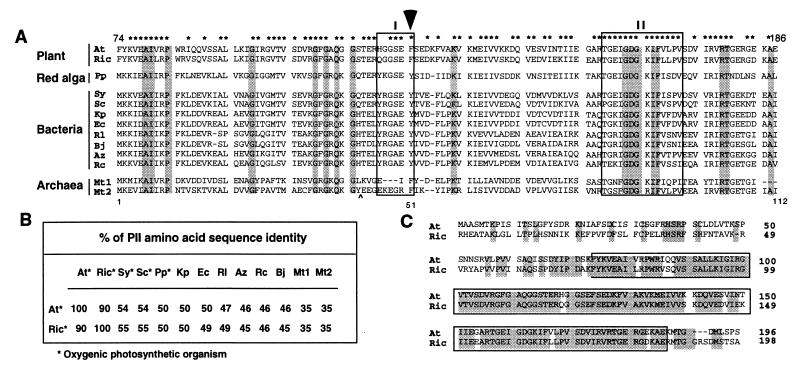
(A) Comparison of the deduced amino acid sequences of plant and microbial PII-like polypeptides. The amino acid residues conserved in all PII-like proteins are shaded. The asterisks (∗) indicate residues that are shared between the plant sequences and any prokaryotic PII-like proteins. Box I and box II refer to PII signature domains I and II. The position of the tyrosine residue that is uridylylated in E. coli is indicated by an arrow. Abbreviations and sequence references: At, A. thaliana; Ric, R. communis; Pp, P. purpurea (16), Sy, Synechocystis sp. PCC 6803 (29); Sc, Synechococcus strain PCC 7942 (30); Kp, K. pneumoniae (31); Ec, E. coli (28); Rl, R. leguminosarum (32); Bj, B. japonicum (33); Az, A. brasilense (34); Rc, R. capsulatus (35); and Mt1 and Mt2, M. thermolithotrophicus PII-like protein 1 and 2 (36). For best alignment, the last two residues GN and a peptide sequence FSANLPEIVDIQKII, which is indicated by “∧”, are deleted in Mt2. The Arabidopsis PII-like protein sequence shown starts with amino acid residue 74 and ends with residue 186. The numbers 1, 51, and 112 indicate the residue position of the E. coli PII protein (product of the glnB gene). (B) The identity of the deduced amino acid sequences of plant and prokaryotic PII proteins. (C) Comparison of the deduced amino acid sequences of plant PII proteins. At, A. thaliana; Ric, R. communis. The identical amino acid residues are shaded. The boxed region corresponds to full-length prokaryotic PII protein.