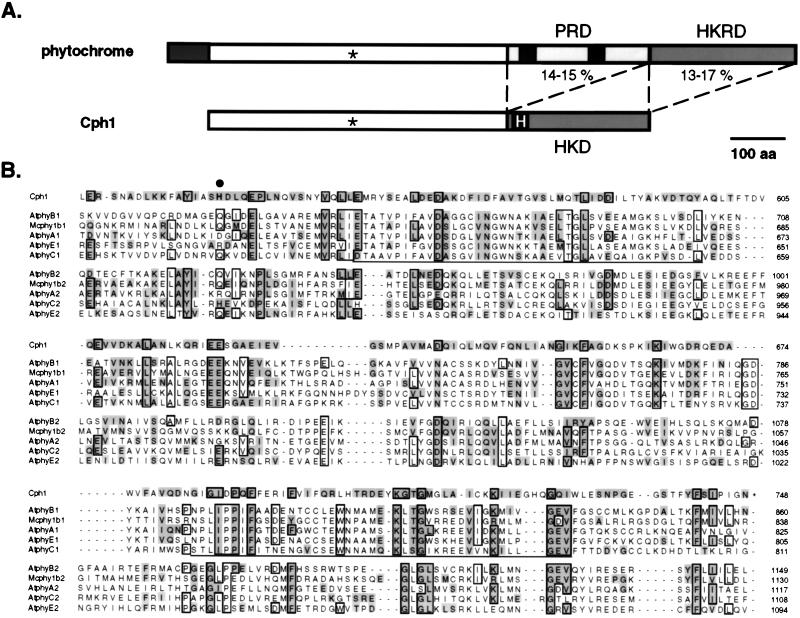
Figure 4.
Duplication of a transmitter module of a prokaryotic phytochrome ancestor: A model for molecular evolution of eukaryotic phytochromes. (A) Structural comparison of eukaryotic phytochromes and Cph1. The conserved cysteine chromophore binding site is marked (∗), and the conserved histidine on the HKD of Cph1 is highlighted. The percent amino acid equivalence between the HKD of Cph1 and both PRD and HKRD of eukaryotic phytochromes is indicated (see Table 1 for details). (B) Multiple sequence alignment of the HKD of Cph1 with the PRD (sequences ending with 1) and HKRD (sequences ending with 2) regions of representative eukaryotic phytochromes including phytochromes from Arabidopsis thaliana (i.e., AtphyA, AtphyB, AtphyC, and AtphyE) and phytochrome from the green alga M. caldariorum. (Mcphy1b). Residues that are identical in six or more sequences are boxed, whereas residues equivalent to those in Cph1 (where I = V, L = M, R = K, S = T = A and D = E = N = Q) are highlighted. The conserved histidine phosphorylation site (H538) of Cph1 is marked with a large dot. The two PAS motifs in eukaryotic phytochromes are underlined. GenBank accession numbers for phytochrome sequences are AtphyA, L21154; AtphyB, X17342; AtphyC, Z32538; AtphyE, X76610; and Mcphy1b, U31284.