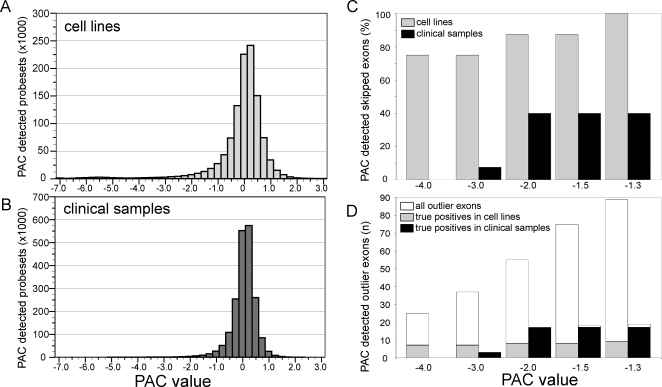
Figure 2. Performance of PAC to detect exon-skipping mutants.
(A) and (B) Total number of PAC-detected outlier probe sets from among 290,000 core probe sets in 12 breast cancer cell lines and in 14 glioblastomas, respectively. (C) Number of skipped exons detected by PAC as a percentage of all eight skipped exons present in the breast cancer cell lines, or as a percentage of the 36 skipped EGFR exons present in the glioblastomas (see Table 1). (D) Total number of outlier exons (true plus false positives) and number of true positive outlier exons detected by PAC among the seven tumor suppressor genes and the EGFR oncogene. True positive outlier exons include all PAC detected skipped exons and two missense mutations (PTEN c.274G>C in CAMA1, MAP2K4 c.551C>G in MDA-MB-134VI).