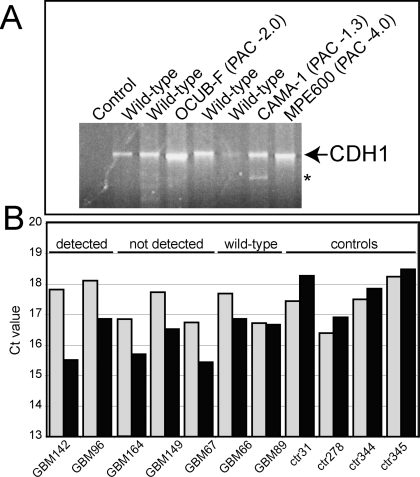
Figure 3. Compromised detection due to heterogeneous transcript expression.
Skipping of CDH1 exon 11 in breast cancer cell line CAMA-1 was only detected at PAC value −1.3, likely due to expression of a second aberrant transcript variant (*) that was detected by conventional RT-PCR. (B) Expression of EGFR transcripts was detected in glioblastoma samples by Real-Time RT-PCR, using primers designed to anneal inside the exon 2–7 deletion region of the EGFRvIII isoform (gray bars) or outside the deletion region (black bars). Differences in Ct values between the two transcript fragments are indicative for EGFRvIII isoform expression levels. All five samples with the EGFRvIII isoform also expressed significant amounts of wild-type EGFR transcripts, likely compromising outlier detection by PAC (indicated by “detected” and “not detected”). Wild-type, samples with normal transcripts; Controls, non-malignant brain specimens.