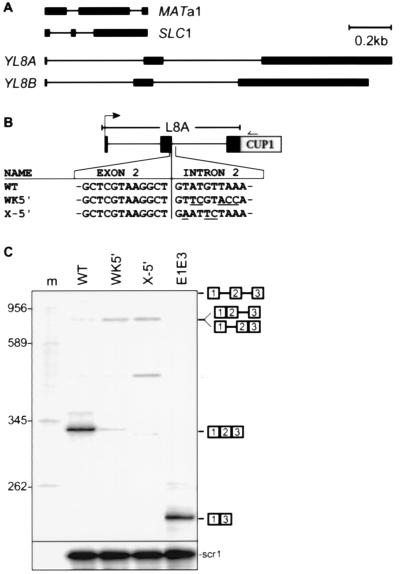
Figure 1.
Splicing phenotype of YL8A mutants suggests an intron definition mechanism. (A) Intron–exon structures of the known multiply interrupted genes in S. cerevisiae. (B) Structure of L8A-CUP1 expression constructs used to test cis-acting factors in YL8A splicing. The sequences shown are at the junction of the second exon and intron. Wild-type (WT) and mutant (WK5′; X-5′) 5′ splice sites were tested (mutant residues are underlined). (C) The 5′ splice site mutations do not induce exon skipping in vivo. Splicing was analyzed by reverse transcription of total cell RNA using a 5′-labeled (32P) primer complementary to CUP1 sequences. Lane m, DNA size markers. E1E3 is a marker for exon skipping expressed from a construct in which exons 1 and 3 are directly fused. Expected products are diagrammed at the right. scr1 is a small cytoplasmic RNA used as an internal control for total RNA amount. A strong RT stop (○) correlates with the 5′ end of snR39, a small nucleolar RNA encoded in intron 2 (24) and could be generated by processing events related to snR39 biosynthesis.