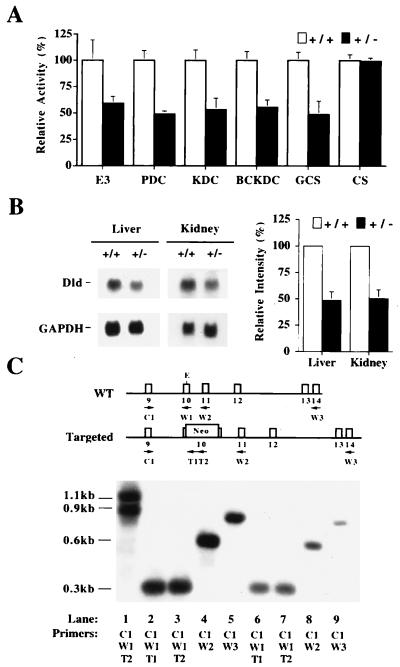
Figure 2.
Analysis of the effect of the Dldtm1mjp. (A) Liver homogenates from Dld+/+ and Dld+/− animals were assayed for E3 activity, each of the E3-dependent complexes and citrate synthase (CS) (n = 4 except for GCS where n = 3). The graph indicates the percentage of activity relative to wild-type levels: E3, 21.0 ± 3.0; PDC, 3.1 ± 0.3; KDC, 5.1 ± 0.4; BCKDC, 0.79 ± 0.05; GCS, 0.88 ± 0.06; CS, 0.17 ± 0.01 (expressed as mean ± SD in milliunits/mg of total protein, except for GCS where milliunits/mg of total mitochondrial protein was used). (B) Northern blot analysis of total RNA from liver and kidney samples of Dld+/+ and Dld+/− animals by using the full-length murine Dld cDNA as a probe. For control of loading, the blots were simultaneously probed with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA probe. A comparison of the normalized intensities with GAPDH is also presented. (C) RT-PCR analysis of total RNA from Dld+/+ and Dld+/− liver samples. The locations of the primer annealing sites are depicted above the blot according to the wild-type and targeted alleles: C1, common primer; W1, W2, and W3, wild-type specific primers for exons 10, 11, and 14, respectively; T1 and T2, mutant specific primers. The amplifications were size-fractionated by gel electrophoresis, blotted, and probed with the murine Dld cDNA. The primers used for each amplification are listed below the lanes. The templates used were as follows. Lanes: 1, Dld+/− genomic DNA; 2–5, Dld+/+ total RNA from liver; 6–9, Dld+/− total RNA from liver. The amplification products of genomic DNA for the wild-type and mutant alleles are 0.9 kb and 1.1 kb, respectively. The amplification products from primer pairs C1/W1, C1/W2, and C1/W3 are 0.3, 0.6, and 0.85 kb, respectively, for the wild-type allele cDNA. For the targeted allele, the amplication products from primer pairs C1/T1 and C1/T2 are 0.4 and 0.5 kb, respectively. E, EcoRI.