Figure 1.
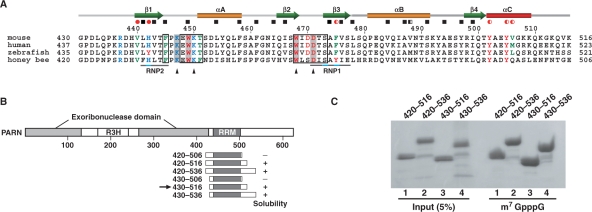
(A) Amino acid sequence alignment of the cap-binding domains of PARNs from different species. The positions of secondary structure elements, as observed in the mouse cap-binding domain, are shown at the top of the alignment. The residues involved in hydrophobic core formation, as determined by GETAREA, are indicated by red circles (V440, H442, F475, Y502, Y505 and V506) and black boxes. Completely and partially buried residues are shown by filled and half-filled symbols, respectively. Residues with obvious chemical shift changes (above the mean value + 1 SD) upon cap binding are boxed. Residues directly involved in cap binding are highlighted in gray. The highly conserved ‘RNP1’ and ‘RNP2’ sequences among the canonical RRMs (52) are aligned with the sequences in the cap-binding domain of PARN and are indicated below the sequences. Positions of amino acid substitutions (see text) are denoted by black triangles. (B) Domain structure of PARN and deletion constructs containing the cap-binding domain of PARN with N- and C-terminal flanking sequences of varying lengths. The RRM region was predicted by the secondary structure elements and is shaded dark gray. Plus and minus signs indicate constructs that yielded soluble protein or inclusion bodies, respectively. The arrow identifies the construct that was selected for the solution structure studies. (C) Pull-down assay of the cap-binding domain with m7G(5′)ppp-Sepharose.