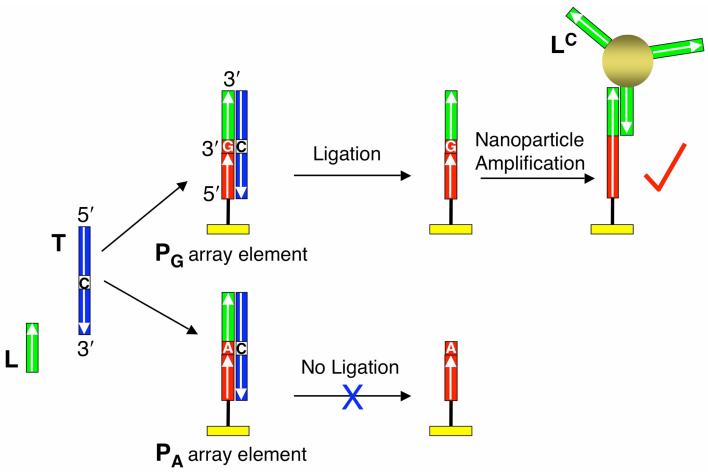
Figure 1.
Schematic representation of the SNP genotyping method based on a combination of surface ligation chemistry and nanoparticle enhanced SPRI. Two array elements with different DNA probes are shown. The array probes (PA and PG) differ only by the last nucleotide at their 3′ ends. When target DNA (T), ligation probe DNA (L) and Taq DNA ligase are simultaneously introduced to the array, surface duplexes form at both array elements. However, ligation only occurs if the duplex is perfectly complementary. After denaturation with 8 M urea, the perfectly matched PG is extended with the L sequence while PA returns to its original state. The presence of L is then detected by the hybridization adsorption of gold nanoparticles modified with oligonucleotides (LC) complementary to L.