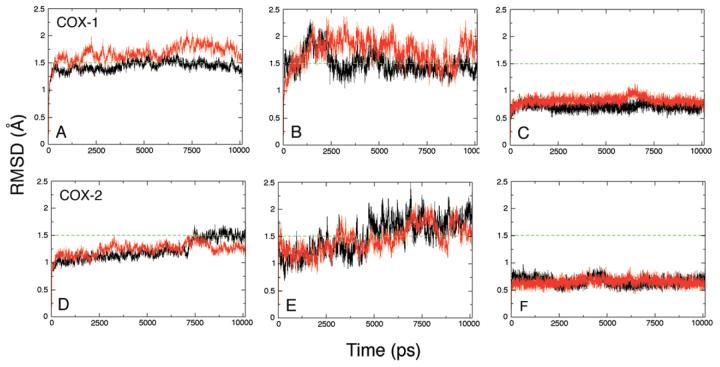
Figure 3.
Root-mean-square deviations for configurations taken at 1 ps intervals from the COX-1 (A, B, C) and COX-2 (D, E, F) simulations, relative to their respective crystal structures, 1DIY and 1CVU. Panels A and D show superposition of backbone atoms (C, CA, N) of monomer A (black) and monomer B (red). Panels B and E show superposition of the backbone atoms of the membrane binding domains alone, residues 73-123. Panels C and F show superposition of the backbone atoms of active site residues, Arg-120, Phe-205, Phe-209, Val-344, Tyr-348, Val-349, Leu-352, Tyr-355, Ile-377, Phe-381, Tyr-385, Trp-387, Ile-523, Gly-526, Ala-527, Ser-530, Gly-533 and Leu-534.