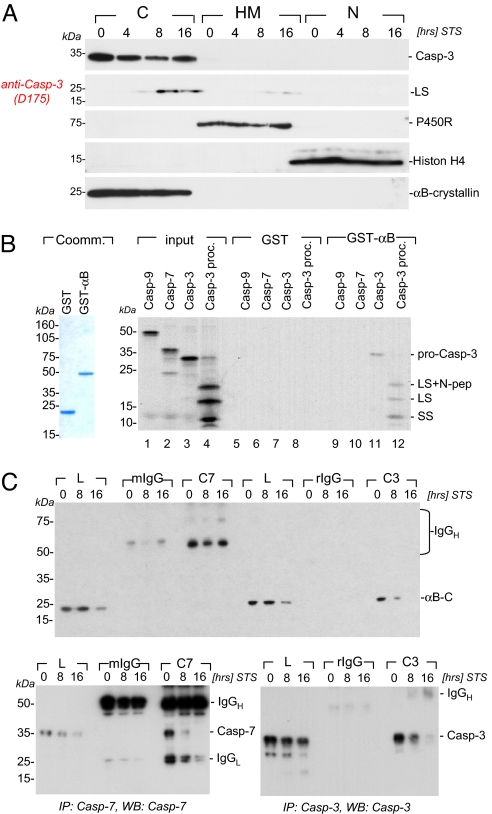
Fig. 3.
αB-crystallin cocompartmentalizes with and selectively binds to the caspase-3 zymogen and its cleavage intermediates. (A) Bcl2L12V5-expressing Ink4a/Arf-deficient astrocytes were treated with STS (1 μM) for the indicated periods of time, and cytosolic (C), heavy membranous (HM), and nuclear (N) compartments were prepared as described in Methods. Besides αB-crystallin, pro- and active caspase-3 species, the migration positions of cytochrome c P450 reductase (P450R), histon H4 and caspase-3 as membrane, nuclear and cytosolic markers are indicated. (B) Affinity-purified GST and GST-αB-crystallin fusion proteins (Coommassie-stained gel for purity assessment) were used in GST pull-down experiments to assess for αB-crystallin:caspase complex formation. GST or GST-αB-crystallin proteins were incubated with [35S]labeled in vitro-translated pro-caspase-9 (Casp-9), pro-caspase-7 (Casp-7), pro-caspase-3 (Casp-3), and pro-caspase-3 preincubated with 20 ng of active caspase-8 (Casp-3proc.) to induce proteolytic processing of the zymogen. A representative autoradiogram with the migration positions of the caspase-3 pro-enzyme and its active subunits (SS, small subunit; LS, large subunit; LS+N pep, large subunit plus N-terminal peptide) is shown. (C) Bcl2L12V5-expressing astrocytes were treated with STS (1 μM) for the indicated periods of time, lysed, and subjected to immunoprecipitation by using monoclonal mouse anti-caspase-7 and rabbit anti-caspase-3 antibodies followed by Western blot analysis under nonreducing conditions for αB-crystallin, caspase-3, and caspase-7. The migration positions of IgG and αB-crystallin, caspase-7, and caspase-3 are indicated.