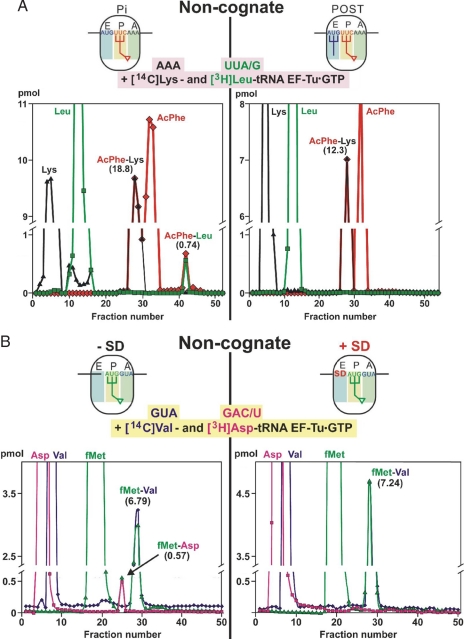
Fig. 2.
Noncognate misincorporation levels. (A) The influence of the E-tRNA: HPLC analysis of dipeptides formed by the addition of a stoichiometric mixture of ternary complexes containing cognate [14C]Lys-tRNA and noncognate [3H]Leu-tRNA to either (i) Pi-state ribosomes (Left) containing AcPhe-tRNA at the P-site or (ii) POST-state ribosomes (Right) carrying AcPhe-tRNA at the P-site and deacylated [32P]tRNAfMet at the E-site, generated via EF-G-dependent translocation. The codons are given above the amino acids, and the specific activity of [3H]Leu-tRNALeu was very high (10,120 dpm/pmol), enabling even very low-level misincorporation of [3H]Leu-tRNALeu to be detected, i.e., the resolution limit was 0.03 pmol for noncognate AcPhe-Leu dipeptide. Puromycin reaction demonstrated a high specificity of >90% for the Pi and POST translocational complexes. (B) The effects of SD on the selection of noncognate aminoacyl-tRNA in the presence of MVF-mRNA. After filling the P-site with f[3H]Met-tRNA (1,670 dpm/pmol), a mixture of ternary complexes was added containing cognate [14C]Val-tRNA (codon GUA; 540 dpm/pmol) and noncognate [3H]Asp-tRNA (GAC/U; 21,750 dpm/pmol). The resolution limit of the noncognate dipeptide fMet-Asp was 0.06 pmol in this HPLC analysis. In the absence of the SD sequence, an error of 7.7% was observed (Left), whereas in its presence, a significant amount of noncognate fMet-Asp is not observed (Right). Each experiment was performed at least three times, a representative run is shown. For further details see Materials and Methods.