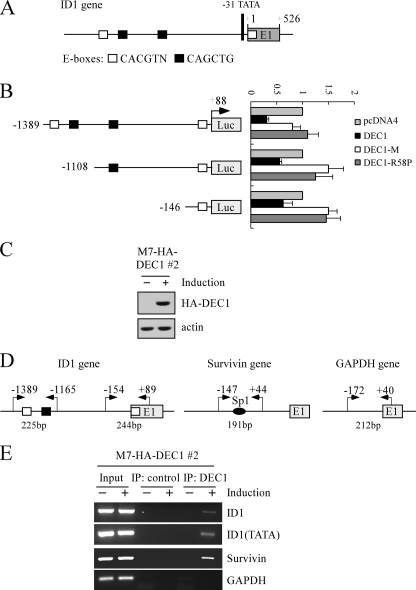
FIGURE 4.
ID1 is a direct target of DEC1. A, schematic presentation of the ID1 genomic structure with the location of the potential DEC1-REs (E-boxes). B, left panel, schematic presentation of three luciferase reporter constructs. See details in the text. Right panel, potential DEC1-REs in the ID1 gene are responsive to wild-type DEC1, but not mutant DEC1-M and DEC1-R58P. The lucifer-ase assay was performed as described under “Experimental Procedures.” C, generation of MCF7 cell lines that inducibly express HA-tagged DEC1. The levels of DEC1 were quantified with anti-HA. D, schematic presentation of the ID1, Survivin, and GAPDH promoters with the locations of potential DEC1-REs and PCR primers used for ChIP assays. E, DEC1 binds to the ID1 promoter in vivo. Upon induction or no induction of DEC1, MCF7 cells were cross-linked with formaldehyde followed by sonication. Chromatin was immunoprecipitated (IP) with anti-HA (HA-DEC1) or a control IgG. DEC1-responsive elements in the ID1 and Survivin genes were amplified by PCR. The binding of DEC1 to the GAPDH was quantified as a nonspecific binding control.