Abstract
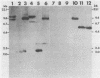
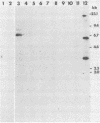
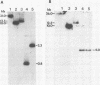
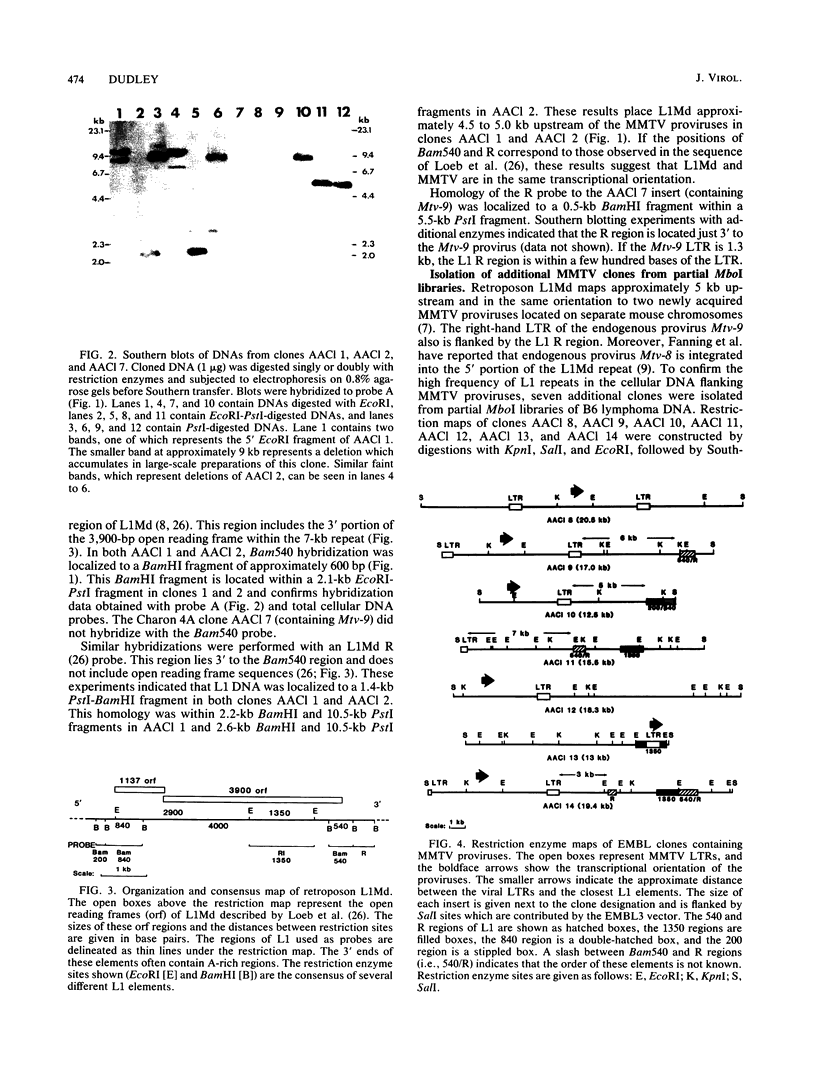
Four Charon 4A clones containing mouse mammary tumor virus (MMTV) proviruses and their cellular flanking sequences were obtained from partial EcoRI libraries of a C57BL/6 T-cell lymphoma with both endogenous and newly acquired MMTV proviruses. The cellular flanking sequences of three of four MMTV proviruses contained DNA homologous to the 3' end of the long interspersed retroposon L1Md. Two of the three proviruses were newly acquired in the lymphoma DNA, and these MMTV proviruses appeared to be 5 kilobases downstream and in the same transcriptional orientation as the L1 sequence. The third provirus was endogenous Mtv-9 and was located less than 500 base pairs from the 3' end of L1. Seven additional clones containing MMTV proviruses were isolated from partial MboI libraries of a B6 T-cell lymphoma. Five of the seven clones contained L1 elements in the cellular DNA flanking MMTV DNA. At least two clones (including one with the Mtv-8 provirus) had multiple L1 copies flanking the MMTV provirus, and one clone contained a single MMTV long terminal repeat directly integrated into a truncated L1 sequence. Although the frequencies of B1 and L1 in random library clones were similar, only one MMTV-containing clone hybridized to the abundant repetitive element B1. These data suggest a nonrandom association between MMTV and L1Md.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. D., Dover G. Organization and evolutionary progress of a dispersed repetitive family of sequences in widely separated rodent genomes. J Mol Biol. 1981 Aug 25;150(4):441–466. doi: 10.1016/0022-2836(81)90374-0. [DOI] [PubMed] [Google Scholar]
- Cohen J. C., Shank P. R., Morris V. L., Cardiff R., Varmus H. E. Integration of the DNA of mouse mammary tumor virus in virus-infected normal and neoplastic tissue of the mouse. Cell. 1979 Feb;16(2):333–345. doi: 10.1016/0092-8674(79)90010-2. [DOI] [PubMed] [Google Scholar]
- Dekaban G. A., Ball J. K. Integration of type B retroviral DNA in virus-induced primary murine thymic lymphomas. J Virol. 1984 Dec;52(3):784–792. doi: 10.1128/jvi.52.3.784-792.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley J. P., Arfsten A., Hsu C. L., Kozak C., Risser R. Molecular cloning and characterization of mouse mammary tumor proviruses from a T-cell lymphoma. J Virol. 1986 Jan;57(1):385–388. doi: 10.1128/jvi.57.1.385-388.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley J. P. Discrete high molecular weight RNA transcribed from the long interspersed repetitive element L1Md. Nucleic Acids Res. 1987 Mar 25;15(6):2581–2592. doi: 10.1093/nar/15.6.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley J., Risser R. Amplification and novel locations of endogenous mouse mammary tumor virus genomes in mouse T-cell lymphomas. J Virol. 1984 Jan;49(1):92–101. doi: 10.1128/jvi.49.1.92-101.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T. G. Characterization of a highly repetitive family of DNA sequences in the mouse. Nucleic Acids Res. 1982 Aug 25;10(16):5003–5013. doi: 10.1093/nar/10.16.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T. G., Morris D. W., Cardiff R. D., Bradshaw H. D., Jr Characterization of an endogenous retrovirus-repetitive DNA chimera in the mouse genome. J Virol. 1985 Mar;53(3):998–1000. doi: 10.1128/jvi.53.3.998-1000.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett D. H., Lister C. K., Kellett E., Finnegan D. J. Transposable elements controlling I-R hybrid dysgenesis in D. melanogaster are similar to mammalian LINEs. Cell. 1986 Dec 26;47(6):1007–1015. doi: 10.1016/0092-8674(86)90815-9. [DOI] [PubMed] [Google Scholar]
- Feinstein S. C., Ross S. R., Yamamoto K. R. Chromosomal position effects determine transcriptional potential of integrated mammary tumor virus DNA. J Mol Biol. 1982 Apr 15;156(3):549–565. doi: 10.1016/0022-2836(82)90266-2. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Furano A. V., Somerville C. C., Tsichlis P. N., D'Ambrosio E. Target sites for the transposition of rat long interspersed repeated DNA elements (LINEs) are not random. Nucleic Acids Res. 1986 May 12;14(9):3717–3727. doi: 10.1093/nar/14.9.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallahan D., Callahan R. Mammary tumorigenesis in feral mice: identification of a new int locus in mouse mammary tumor virus (Czech II)-induced mammary tumors. J Virol. 1987 Jan;61(1):66–74. doi: 10.1128/jvi.61.1.66-74.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray D. A., Chan E. C., MacInnes J. I., Morris V. L. Restriction endonuclease map of endogenous mouse mammary tumor virus loci in GR, DBA, and NFS mice. Virology. 1986 Jan 15;148(1):237–242. doi: 10.1016/0042-6822(86)90421-6. [DOI] [PubMed] [Google Scholar]
- Green P. L., Lamph W. W., Dudley J., Arfsten A., Risser R., Lanier L. L., Warner N. L., Tung J. S., Scheid M. P. Phenotypic variation in clonal Abelson virus lymphoma cells. J Immunol. 1985 Feb;134(2):1268–1275. [PubMed] [Google Scholar]
- Groner B., Buetti E., Diggelmann H., Hynes N. E. Characterization of endogenous and exogenous mouse mammary tumor virus proviral DNA with site-specific molecular clones. J Virol. 1980 Dec;36(3):734–745. doi: 10.1128/jvi.36.3.734-745.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes S. R., Jelinek W. R. Low molecular weight RNAs transcribed in vitro by RNA polymerase III from Alu-type dispersed repeats in Chinese hamster DNA are also found in vivo. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6130–6134. doi: 10.1073/pnas.78.10.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers J., Bentvelzen P. Interaction between viral and genetic factors in murine mammary cancer. Adv Cancer Res. 1978;26:143–195. doi: 10.1016/s0065-230x(08)60087-1. [DOI] [PubMed] [Google Scholar]
- Hynes N. E., Kennedy N., Rahmsdorf U., Groner B. Hormone-responsive expression of an endogenous proviral gene of mouse mammary tumor virus after molecular cloning and gene transfer into cultured cells. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2038–2042. doi: 10.1073/pnas.78.4.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins N. A., Copeland N. G., Taylor B. A., Lee B. K. Dilute (d) coat colour mutation of DBA/2J mice is associated with the site of integration of an ecotropic MuLV genome. Nature. 1981 Oct 1;293(5831):370–374. doi: 10.1038/293370a0. [DOI] [PubMed] [Google Scholar]
- Kennedy N., Knedlitschek G., Groner B., Hynes N. E., Herrlich P., Michalides R., van Ooyen A. J. Long terminal repeats of endogenous mouse mammary tumour virus contain a long open reading frame which extends into adjacent sequences. Nature. 1982 Feb 18;295(5850):622–624. doi: 10.1038/295622a0. [DOI] [PubMed] [Google Scholar]
- Kramerov D. A., Grigoryan A. A., Ryskov A. P., Georgiev G. P. Long double-stranded sequences (dsRNA-B) of nuclear pre-mRNA consist of a few highly abundant classes of sequences: evidence from DNA cloning experiments. Nucleic Acids Res. 1979 Feb;6(2):697–713. doi: 10.1093/nar/6.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krayev A. S., Kramerov D. A., Skryabin K. G., Ryskov A. P., Bayev A. A., Georgiev G. P. The nucleotide sequence of the ubiquitous repetitive DNA sequence B1 complementary to the most abundant class of mouse fold-back RNA. Nucleic Acids Res. 1980 Mar 25;8(6):1201–1215. doi: 10.1093/nar/8.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon B. S., Weissman S. M. Mouse mammary tumor virus-related sequences in mouse lymphocytes are inducible by 12-O-tetradecanoyl phorbol-13-acetate. J Virol. 1984 Dec;52(3):1000–1004. doi: 10.1128/jvi.52.3.1000-1004.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb D. D., Padgett R. W., Hardies S. C., Shehee W. R., Comer M. B., Edgell M. H., Hutchison C. A., 3rd The sequence of a large L1Md element reveals a tandemly repeated 5' end and several features found in retrotransposons. Mol Cell Biol. 1986 Jan;6(1):168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981 Jan 22;289(5795):253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- Martin S. L., Voliva C. F., Burton F. H., Edgell M. H., Hutchison C. A., 3rd A large interspersed repeat found in mouse DNA contains a long open reading frame that evolves as if it encodes a protein. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2308–2312. doi: 10.1073/pnas.81.8.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalides R., Wagenaar E., Hilkens J., Hilgers J., Groner B., Hynes N. E. Acquisition of proviral DNA of mouse mammary tumor virus in thymic leukemia cells from GR mice. J Virol. 1982 Sep;43(3):819–829. doi: 10.1128/jvi.43.3.819-829.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalides R., Wagenaar E., Weijers P. Rearrangements in the long terminal repeat of extra mouse mammary tumor proviruses in T-cell leukemias of mouse strain GR result in a novel enhancer-like structure. Mol Cell Biol. 1985 Apr;5(4):823–830. doi: 10.1128/mcb.5.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R., Varmus H. E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982 Nov;31(1):99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- Nusse R., van Ooyen A., Cox D., Fung Y. K., Varmus H. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature. 1984 Jan 12;307(5947):131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- Peters G., Brookes S., Smith R., Dickson C. Tumorigenesis by mouse mammary tumor virus: evidence for a common region for provirus integration in mammary tumors. Cell. 1983 Jun;33(2):369–377. doi: 10.1016/0092-8674(83)90418-x. [DOI] [PubMed] [Google Scholar]
- Peters G., Lee A. E., Dickson C. Concerted activation of two potential proto-oncogenes in carcinomas induced by mouse mammary tumour virus. Nature. 1986 Apr 17;320(6063):628–631. doi: 10.1038/320628a0. [DOI] [PubMed] [Google Scholar]
- Peters G., Placzek M., Brookes S., Kozak C., Smith R., Dickson C. Characterization, chromosome assignment, and segregation analysis of endogenous proviral units of mouse mammary tumor virus. J Virol. 1986 Sep;59(3):535–544. doi: 10.1128/jvi.59.3.535-544.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson D. O., Kriz K. G., Marich J. E., Toohey M. G. Sequence organization and molecular cloning of mouse mammary tumor virus DNA endogenous to C57BL/6 mice. J Virol. 1985 May;54(2):525–531. doi: 10.1128/jvi.54.2.525-531.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J. H. The origin and evolution of retroposons. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Voliva C. F., Jahn C. L., Comer M. B., Hutchison C. A., 3rd, Edgell M. H. The L1Md long interspersed repeat family in the mouse: almost all examples are truncated at one end. Nucleic Acids Res. 1983 Dec 20;11(24):8847–8859. doi: 10.1093/nar/11.24.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voliva C. F., Martin S. L., Hutchison C. A., 3rd, Edgell M. H. Dispersal process associated with the L1 family of interspersed repetitive DNA sequences. J Mol Biol. 1984 Oct 5;178(4):795–813. doi: 10.1016/0022-2836(84)90312-7. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Wilson R., Storb U. Association of two different repetitive DNA elements near immunoglobulin light chain genes. Nucleic Acids Res. 1983 Mar 25;11(6):1803–1817. doi: 10.1093/nar/11.6.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]