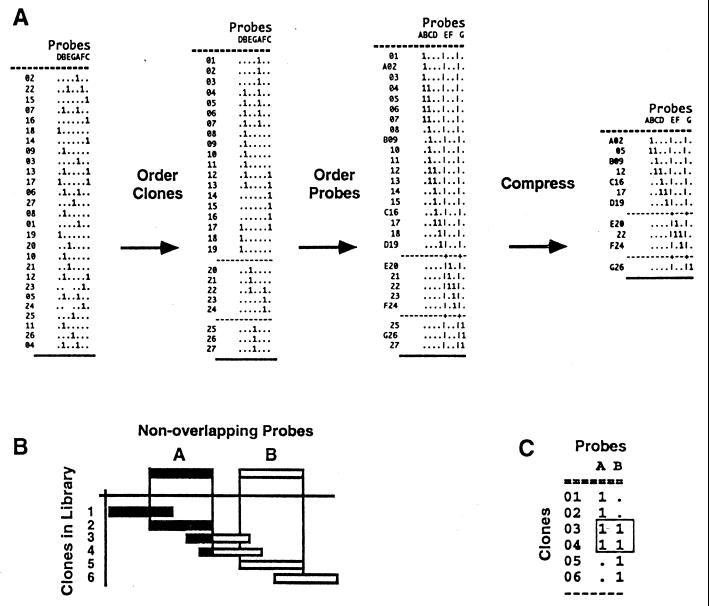
Figure 1.
The two-way complementary ordering process and compression of a binary clone/probe hybridization matrix. A schematic representation of the ordering process of DNA/DNA hybridization data into a physical map. All clones present in the library are indexed by rows (in the example clones are indicated by numbers), and nonoverlapping probes are listed across the columns (in the example probes are indicated by letters). The S, R, and O designations were appended to the clone names (17). Probes are clones selected from the chromosome-specific clone collection (846 probes) and hybridized to filters containing the entire library. In the binary matrix, presence or absence of hybridization of each clone to each probe is indicated by 1 or “.”, respectively. Clones were ordered by simulated annealing (18) or random cost (14) algorithms, and contigs are represented by blocks of linked clones separated by lines that indicate contig boundaries. To order nonoverlapping probes, the clone/probe hybridization matrix was transposed and reordered in the computer. For both algorithms the number of differences between successive clones (probes) down the rows (columns) is computed and summed to form the total linking distance D. This total linking distance is minimized by randomly permuting the rows (or columns) and has been shown to produce the right ordering of clones for a large number of probes (17). This two-way ordering process establishes complementary minimal and redundant orders of probe and clone collections, respectively. Compression of physical maps by elimination of redundant clones results in a minimal map that retains all of the probes and one overlapping clone linking a pair of probes. A schematic of the ordering concept of nonoverlapping probes by overlapping clones is shown in B and the binary information that orders probes in C.